Back
Back Sanders 3rd Edition
Sanders 3rd Edition Ch. 20 - Population Genetics and Evolution at the Population, Species, and Molecular Levels
Ch. 20 - Population Genetics and Evolution at the Population, Species, and Molecular LevelsProblem D.1
Why might mitochondrial, Y chromosome, and autosomal DNA provide different perspectives on our evolutionary past, for example, with respect to our relationship with Neanderthals?
Problem 1a
Compare and contrast the terms in each of the following pairs:
Population and gene pool
Problem 1b
Compare and contrast the terms in each of the following pairs:
Random mating and Inbreeding
Problem 1c
Compare and contrast the terms in each of the following pairs:
Natural selection and Genetic drift
Problem 1d
Compare and contrast the terms in each of the following pairs:
A polymorphic trait and a polymorphic gene
Problem 1e
Compare and contrast the terms in each of the following pairs:
Founder effect and Genetic bottleneck
- In a population, what is the consequence of inbreeding? Does inbreeding change allele frequencies? What is the effect of inbreeding with regard to rare recessive alleles in a population?
Problem 2
Problem D.2
What insights have analyses of human mitochondrial DNA provided into our recent evolutionary past?
Problem 3
Identify and describe the evolutionary forces that can cause allele frequencies to change from one generation to the next.
Problem D.3
What lines of evidence support the hypothesis that modern humans evolved in Africa and then subsequently migrated throughout the globe?
Problem 4
Describe how natural selection can produce balanced polymorphism of allele frequencies through selection that favors heterozygotes.
Problem D.4
Discuss how both gains and losses of regulatory elements may lead to human-specific traits.
Problem 5
Thinking creatively about evolutionary mechanisms, identify at least two schemes that could generate allelic polymorphism in a population. Do not include the processes described in the answer to Problem 4.
Problem D.5
How do copy-number variants arise? Do they account for more polymorphism than SNPs within the human population?
Problem 6
Genetic drift, an evolutionary process affecting all populations, can have a significant effect in small populations, even though its effect is negligible in large populations. Explain why this is the case.
Problem D.6
Consider possible societal and ethical dilemmas that might arise if we currently shared the planet with another hominin.
- Over the course of many generations in a small population, what effect does random genetic drift have on allele frequencies?
Problem 7
Problem D.7
Carl Linnaeus, the 18th-century botanist who laid the foundation for the modern system of taxonomic nomenclature, placed chimpanzees and humans in the same genus. Discuss the merits of this classification.
- Catastrophic events such as loss of habitat, famine, or overhunting can push species to the brink of extinction and result in a genetic bottleneck. What happens to allele frequencies in a species that experiences a near-extinction event, and what is expected to happen to allele frequencies if the species recovers from near extinction?
Problem 8
Problem D.8
Describe how selection at a locus can result in a loss of polymorphism surrounding the locus.
- George Udny Yule was wrong in suggesting that an autosomal dominant trait like brachydactyly will increase in frequency in populations. Explain why Yule was incorrect.
Problem 9
Problem D.9
How can ancient DNA provide insight into past migrations that analyses of extant human genomes fail to uncover?
Problem D.10
Denisovans are known from bones found in Denisova Cave in the Altai Mountains in Siberia, but traces of their DNA are found in Australians and Melanesians, whose ancestors likely migrated across Asia much farther to the south. How can these geographic differences be reconciled?
Problem 10a
The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of the recessive allele.
Problem 10b
The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of the dominant allele.
Problem 10c
The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of each genotype.
Problem 11
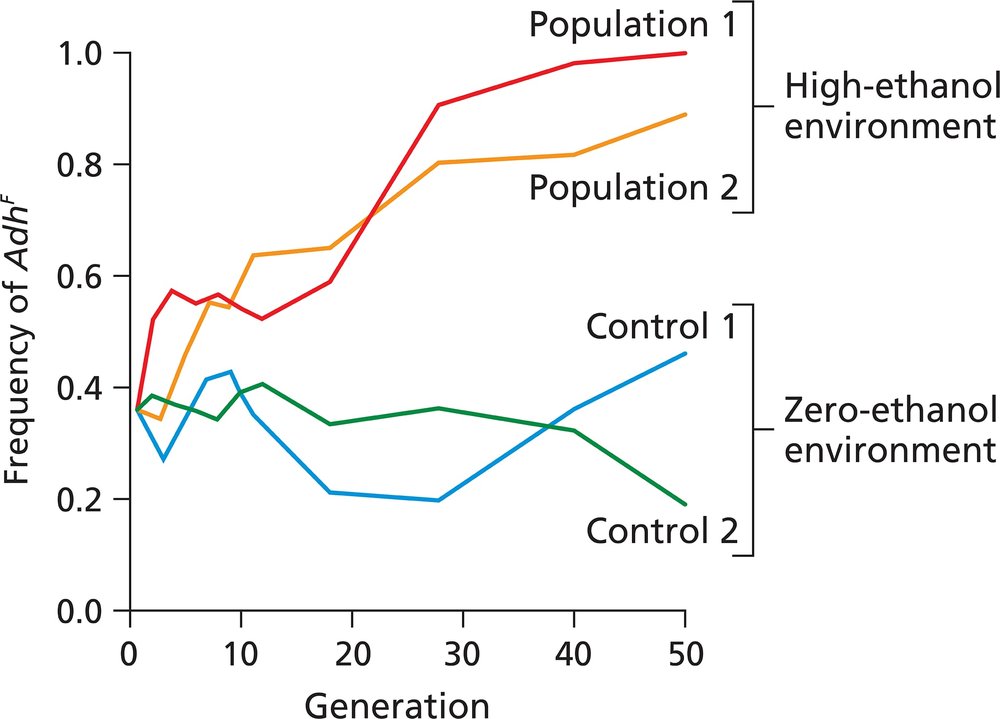
The figure illustrates the effect of an ethanol-rich and an ethanol-free environment on the frequency of the Drosophila AdhF allele in four populations in a 50-generation laboratory experiment. Population 1 and population 2 were reared for 50 generations in a high-ethanol environment, while control 1 and control 2 populations were reared for 50 generations in a zero-ethanol environment. Describe the effect of each environment on the populations, and state any conclusions you can reach about the role of any of the evolutionary processes in producing these effects.
Problem D.11
In Island Melanesia and Polynesia, most mtDNA haplotypes are of Asian ancestry, whereas Y chromosome haplotypes are predominantly New Guinean. Provide a hypothesis for this sex-biased distribution.
- Biologists have proposed that the use of antibiotics to treat human infectious disease has played a role in the evolution of widespread antibiotic resistance in several bacterial species, including Staphylococcus aureus and the bacteria causing gonorrhea, tuberculosis, and other infectious diseases. Explain how the evolutionary mechanisms mutation and natural selection may have contributed to the development of antibiotic resistance.
Problem 12
Problem D.12
A 9-bp deletion in the mitochondrial genome between the gene for cytochrome oxidase subunit II and the gene for tRNAᴸʸˢ is a common polymorphism among Polynesians and also in a population of Taiwanese natives. The frequency of the polymorphism varies between populations: The highest frequency is seen in the Maoris of New Zealand (98%), lower levels are seen in eastern Polynesia (80%) and western Polynesia (89%), and the lowest level is seen in the Taiwanese population. What do these frequencies tell us about the settlement of the Pacific by the ancestors of the present-day Polynesians?