
 Sanders 3rd Edition
Sanders 3rd Edition Ch. 20 - Population Genetics and Evolution at the Population, Species, and Molecular Levels
Ch. 20 - Population Genetics and Evolution at the Population, Species, and Molecular Levels Problem 11
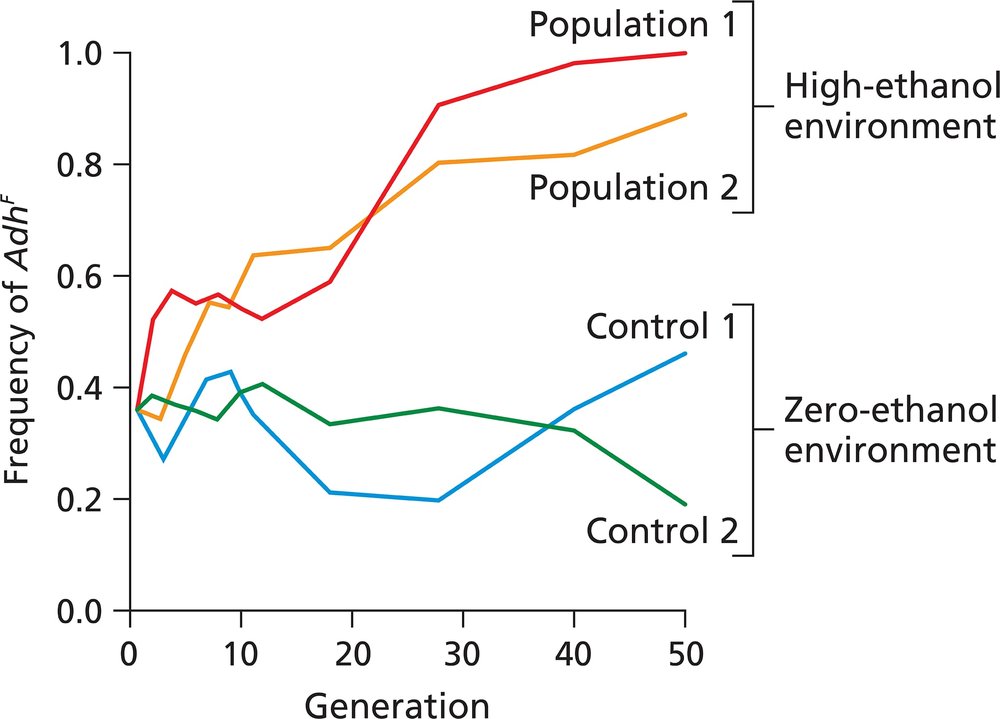
Problem 11The figure illustrates the effect of an ethanol-rich and an ethanol-free environment on the frequency of the Drosophila AdhF allele in four populations in a 50-generation laboratory experiment. Population 1 and population 2 were reared for 50 generations in a high-ethanol environment, while control 1 and control 2 populations were reared for 50 generations in a zero-ethanol environment. Describe the effect of each environment on the populations, and state any conclusions you can reach about the role of any of the evolutionary processes in producing these effects.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Natural Selection
Allele Frequency and Genetic Variation

Role of Environmental Pressure in Evolution

The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of the recessive allele.
The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of the dominant allele.
The ability to taste the bitter compound phenylthiocarbamide (PTC) is an autosomal dominant trait. The inability to taste PTC is a recessive condition. In a sample of 500 people, 360 have the ability to taste PTC and 140 do not. Calculate the frequency of each genotype.
In Island Melanesia and Polynesia, most mtDNA haplotypes are of Asian ancestry, whereas Y chromosome haplotypes are predominantly New Guinean. Provide a hypothesis for this sex-biased distribution.
A 9-bp deletion in the mitochondrial genome between the gene for cytochrome oxidase subunit II and the gene for tRNAᴸʸˢ is a common polymorphism among Polynesians and also in a population of Taiwanese natives. The frequency of the polymorphism varies between populations: The highest frequency is seen in the Maoris of New Zealand (98%), lower levels are seen in eastern Polynesia (80%) and western Polynesia (89%), and the lowest level is seen in the Taiwanese population. What do these frequencies tell us about the settlement of the Pacific by the ancestors of the present-day Polynesians?