Gene G recombines with gene T at a frequency of 7%, and gene G recombines with gene R at a frequency of 4%.
Draw two possible genetic maps for these three genes, and identify the recombination frequencies predicted for each map.

 Verified step by step guidance
Verified step by step guidance
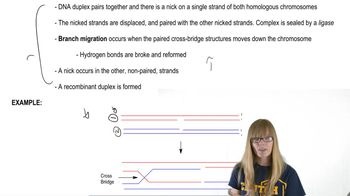
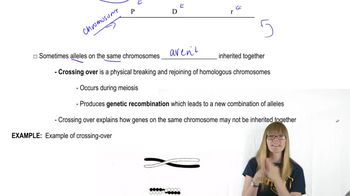
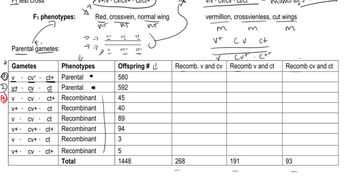
Gene G recombines with gene T at a frequency of 7%, and gene G recombines with gene R at a frequency of 4%.
Draw two possible genetic maps for these three genes, and identify the recombination frequencies predicted for each map.
In Drosophila, the map positions of genes are given in map units numbering from one end of a chromosome to the other. The X chromosome of Drosophila is 66 m.u. long. The X-linked gene for body color—with two alleles, y⁺ for gray body and y for yellow body—resides at one end of the chromosome at map position 0.0. A nearby locus for eye color, with alleles w⁺ for red eye and w for white eye, is located at map position 1.5. A third X-linked gene, controlling bristle form, with f⁺ for normal bristles and f for forked bristles, is located at map position 56.7. At each locus the wild-type allele is dominant over the mutant allele.
Explain how each of the predicted progeny classes is produced.
Genes A, B, and C are linked on a chromosome and found in the order A–B–C. Genes A and B recombine with a frequency of 8%, and genes B and C recombine at a frequency of 24%. For the cross a⁺b⁺c/abc⁺ × abc/abc, predict the frequency of progeny genotypes. Assume interference is zero.
Genes A, B, C, D, and E are linked on a chromosome and occur in the order given.
The test cross Ae/aE x ae/ae indicates the genes recombine with a frequency of 28%. If 1000 progeny are produced by this test cross, determine the number of progeny in each outcome class.
Genes A, B, C, D, and E are linked on a chromosome and occur in the order given.
Previous genetic linkage crosses have determined that recombination frequencies are 6% for genes A and B, 4% for genes B and C, 10% for genes C and D, and 11% for genes D and E. The sum of these frequencies between genes A and E is 31%. Why does the recombination distance between these genes as determined by adding the intervals between adjacent linked genes differ from the distance determined by the test cross?
Syntenic genes can assort independently. Explain this observation.