A trihybrid cross involves examining the inheritance of three different traits simultaneously, similar to a dihybrid cross but with an additional trait. To analyze the offspring from such a cross, a Punnett square can be utilized to determine the genotypes and phenotypes of the resulting offspring. The principles of Mendelian inheritance apply, particularly when dealing with unlinked genes, which segregate independently. However, for linked genes located on the same chromosome, the recombination frequencies of the offspring can provide insights into their genetic locations.
In a practical example using fruit flies, three traits are considered: eye color, wing veins, and wing shape. The wild type allele for eye color is represented by red eyes (denoted as +), while the mutant allele is vermilion. For wing veins, the presence of a cross vein indicates the wild type, while the absence indicates the mutant. Wing shape is classified as normal for the wild type and cut short for the mutant. The parental generation consists of one individual homozygous for the wild type eye color and mutant for the other two traits, crossed with another individual that is mutant for eye color and homozygous wild type for the other traits.
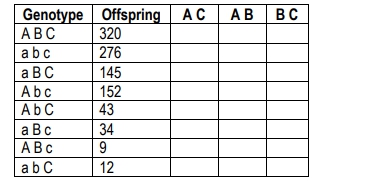
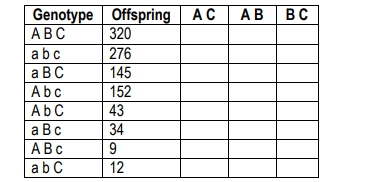
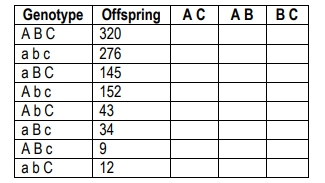
The resulting F1 generation will be heterozygous for all traits, allowing for the tracking of allele transmission. When performing a test cross with a homozygous recessive individual, the phenotypes of the offspring can be analyzed to determine which alleles are parental and which are recombinant. Parental gametes are those that match the original parental phenotypes, while recombinant gametes exhibit new combinations of traits due to crossing over during meiosis.
To identify which alleles have recombined, each offspring's genotype is compared to the parental genotypes. The differences indicate which alleles have undergone recombination. The recombination frequency can then be calculated using the formula:
\[ RF = \frac{\text{Number of recombinant offspring}}{\text{Total offspring}} \]
For example, if the recombination frequencies for the traits are calculated as follows: 18.5% for the first pair of genes, 13.2% for the second, and 6.4% for the third, it indicates that all three genes are linked since their recombination frequencies are less than 50%. This information can be used to create a chromosome map, placing the genes in order based on their recombination frequencies. The gene with the highest frequency is placed farthest apart, while the others are positioned based on their relative distances.
It is crucial to account for potential double crossovers, which can affect the recombination frequency calculations. By recognizing that double crossovers may lead to discrepancies in expected distances, adjustments can be made to ensure accurate mapping. Understanding these concepts is essential for success in genetics, particularly in predicting inheritance patterns and mapping gene locations.