Back
Back- You have discovered a new species of archaea from a hot spring in Yellowstone National Park. After growing a pure culture of this organism, what strategy might you employ to sequence its genome?
Problem 1
Problem 1b
You have discovered a new species of archaea from a hot spring in Yellowstone National Park. How would your strategy change if you were unable to grow the strain in culture?
Problem 2a
Repetitive DNA poses problems for genome sequencing. Why is this so?
Problem 2b
Repetitive DNA poses problems for genome sequencing. What types of repetitive DNA are most problematic?
Problem 2c
Repetitive DNA poses problems for genome sequencing. What strategies can be employed to overcome these problems?
Problem 3a
When the whole-genome shotgun sequence of the Drosophila genome was assembled, it comprised 134 scaffolds made up of 1636 contigs. Why were there so many more contigs than scaffolds?
Problem 3b
When the whole-genome shotgun sequence of the Drosophila genome was assembled, it comprised 134 scaffolds made up of 1636 contigs. What is the difference between physical and sequence gaps?
Problem 3c
When the whole-genome shotgun sequence of the Drosophila genome was assembled, it comprised 134 scaffolds made up of 1636 contigs. How can physical gaps be closed?
Problem 3d
When the whole-genome shotgun sequence of the Drosophila genome was assembled, it comprised 134 scaffolds made up of 1636 contigs.
How can sequence gaps be closed?
Problem 4
How do cDNA sequences facilitate gene annotation? Describe how the use of full-length cDNAs facilitates discovery of alternative splicing.
Problem 5
How do comparisons between genomes of related species help refine gene annotation?
Problem 6a
You are designing algorithms for the bioinformatic prediction of gene sequences. How might algorithms differ for predicting genes in bacterial versus eukaryotic genomic sequence?
Problem 7
You have sequenced a 100-kb region of the Bacillus anthracis genome (the bacterium that causes anthrax) and a 100-kb region from the Gorilla gorilla genome. What differences and similarities might you expect to see in the annotation of the sequences, for example, in the number of genes, gene structure, regulatory sequences, and repetitive DNA?
Problem B.7
Diseases and conditions on the RUSP list are tested on every newborn infant, and if the baby has one of the conditions, the parents are immediately informed. What kind of information and counseling should be provided to the parents along with the diagnosis?
Problem 8
You have just obtained 100 kb of genomic sequence from an as-yet-unsequenced mammalian genome. What are three methods you might use to identify potential genes in the 100 kb? What are the advantages and limitations of each method?
Problem 9
The human genome contains a large number of pseudogenes. How would you distinguish whether a particular sequence encodes a gene or a pseudogene? How do pseudogenes arise?
Problem 10
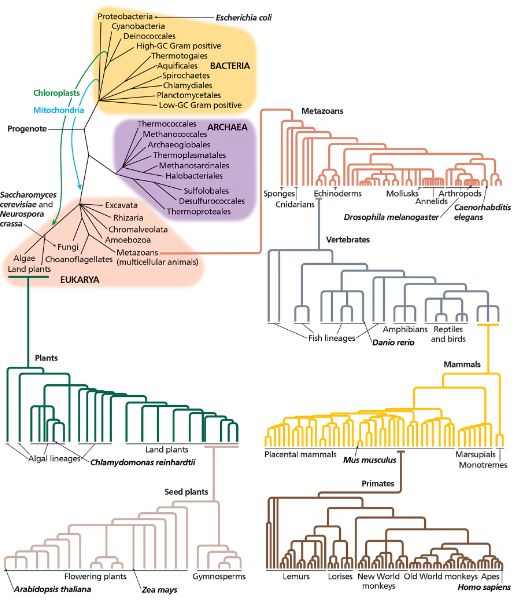
Based on the tree of life in the following figure (Figure 16.12), would you expect human proteins to be more similar to fungal proteins or to plant proteins? Would you expect plant proteins to be more similar to fungal proteins or to human proteins?
Problem 11
When comparing genes from two sequenced genomes, how does one determine whether two genes are orthologous? What pitfalls arise when one or both of the genomes are not sequenced?
Problem 12
What is a reference genome? How can it be used to survey genetic variation within a species?
Problem 13
The two-hybrid method facilitates the discovery of protein–protein interactions. How does this technique work? Can you think of reasons for obtaining a false-positive result, that is, where the proteins encoded by two clones interact in the two-hybrid system but do not interact in the organism in which they naturally occur? Can you think of reasons you might obtain a false-negative result, in which the two proteins interact in vivo but fail to interact in the two-hybrid system?
Problem B.13
Imagine yourself in the same position as Kristen Powers, faced with the decision of whether or not to undergo a genetic test that will discover if you have inherited Huntington disease. List five life decisions or choices that you think are likely to be affected by the results of the genetic test. Do you think you would make the same choice to test that Kristen made? Why or why not?
Problem 14
Go to http://blast.ncbi.nlm.nih.gov/Blast.cgi and follow the links to nucleotide BLAST. Type in the sequence below; it is broken up into codons to make it easier to copy.
5' ATG TTC GTC AAT CAG CAC CTT TGT GGT TCT CAC CTC GTT GAA GCTTTG TAC CTT GTT TGC GGT GAA CGT GGT TTC TTC TAC ACT CCT AAG ACT TAA 3'
As you will note on the BLAST page, there are several options for tailoring your query to obtain the most relevant information. Some are related to which sequences to search in the database. For example, the search can be limited taxonomically (e.g., restricted to mammals) or by the type of sequences in the database (e.g., cDNA or genomic). For our search, we will use the broadest database, the 'Nucleotide collection (nr/nt).' This is the nonredundant (nr) database of all nucleotide data (nt) in GenBank and can be selected in the 'Database' dialogue box. Other parameters can also be adjusted to make the search more or less sensitive to mismatches or gaps. For our purposes, we will use the default setting, which is automatically presented. Press 'BLAST' to search. What can you say about the DNA sequence?
Problem B.14e
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information:
The duration of treatment
Problem B.14d
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information:
The recommended treatment for those with the condition.
Problem B.14a
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information: The frequency of the condition in newborn infants (note any populations in which the condition is more frequent)
Problem B.14b
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information:
The defect that characterizes the condition.
Problem B.14f
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information:
The anticipated outcome if treatment is applied
Problem B.14c
Select one of the hereditary conditions from either the RUSP core conditions list or the RUSP list of secondary conditions and do some online research to find the following information:
The symptoms and consequences of the condition if it is not treated.
Problem 15
In the course of the Drosophila melanogaster genome project, the following genomic DNA sequences were obtained. Try to assemble the sequences into a single contig.
5' TTCCAGAACCGGCGAATGAAGCTGAAGAAG 3'
5' GAGCGGCAGATCAAGATCTGGTTCCAGAAC 3'
5' TGATCTGCCGCTCCGTCAGGCATAGCGCGT 3'
5' GGAGAATCGAGATGGCGCACGCGCTATGCC 3'
5' GGAGAATCGAGATGGCGCACGCGCTATGCC 3'
5' CCATCTCGATTCTCCGTCTGCGGGTCAGAT 3'
Go to the URL provided in Problem 14, and using the sequence you have just assembled, perform a blastn search in the 'Nucleotide collection (nr/nt)' database. Does the search produce sequences similar to your assembled sequence, and if so, what are they? Can you tell if your sequence is transcribed, and if it represents protein-coding sequence? Perform a tblastx search, first choosing the 'Nucleotide collection (nr/nt)' database and then limiting the search to human sequences by typing Homo sapiens in the organism box. Are homologous sequences found in the human genome? Annotate the assembled sequence.
Problem 16a
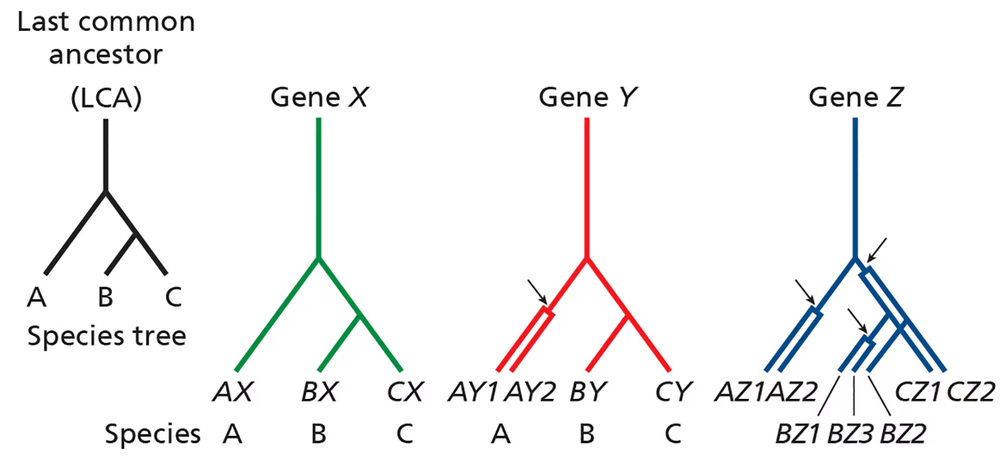
Consider the phylogenetic trees below pertaining to three related species (A, B, and C) that share a common ancestor (last common ancestor, or LCA). The lineage leading to species A diverges before the divergence of species B and C.
For gene X, no gene duplications have occurred in any lineage, and each gene X is derived from the ancestral gene X via speciation events. Are genes AX, BX, and CX orthologous, paralogous, or homologous?