From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Separate the chromosomes and chromatids as though mitotic anaphase and telophase have taken place.

From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Are there any alternative alignments of the chromosomes for this cell-division stage? Explain.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Chromatids and Chromosomes
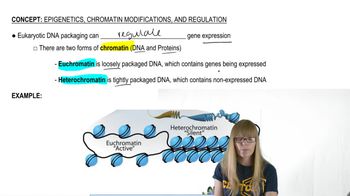
Homologous Chromosomes
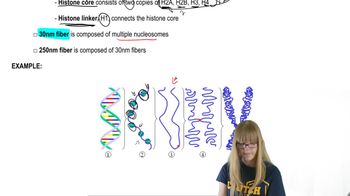
Cell Division and Alignment

From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. What are the genotypes of the daughter cells?
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Align the chromosomes as they might appear at metaphase I of meiosis.
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Separate the chromosomes as though meiotic anaphase I and telophase I have taken place.
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Align the chromosomes of each daughter cell as they might appear in metaphase II of meiosis.
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Are there any alternative alignments of the chromosomes for this cell-division stage? Explain.
