Back
BackProblem 10
Describe the differences between genetic complementation and recombination as they relate to the detection of wild-type lysis by a mutant bacteriophage.
Problem 11
Among the mechanisms of gene transfer in bacteria, which one is capable of transferring the largest chromosome segment from donor to recipient? Which process generally transfers the smallest donor segments to the recipient? Explain your reasoning for both answers.
Problem 12
What is lateral gene transfer? How might it take place between two bacterial cells?
Problem 13
Lateral gene transfer is thought to have played a major role in the evolution of bacterial genomes. Describe the impact of LGT on bacterial genome evolution.
Problem 14
Seven deletion mutations (1 to 7 in the table below) are tested for their ability to form wild-type recombinants with five point mutations (a to e). The symbol "+" indicates that wild-type recombination occurs, and "-" indicates that wild types are not formed. Use the data to construct a genetic map of the order of point mutations, and indicate the segment deleted by each deletion mutation.

Problem 15a
A 2013 CDC report identified the practice of routinely adding antibiotic compounds to animal feed as a major culprit in the rapid increase in the number of antibiotic-resistant strains. Agricultural practice in recent decades has encouraged the addition of antibiotics to animal feed to promote growth rather than to treat disease.
Speculate about the process by which feeding antibiotics to animals such as cattle might lead to an increase in the number of antibiotic-resistant strains of bacteria.
Problem 15b
A 2013 CDC report identified the practice of routinely adding antibiotic compounds to animal feed as a major culprit in the rapid increase in the number of antibiotic-resistant strains. Agricultural practice in recent decades has encouraged the addition of antibiotics to animal feed to promote growth rather than to treat disease.
How might the increase in antibiotic-resistant strains of bacteria in cattle be a threat to human health?
Problem 16a
Hfr strains that differ in integrated F factor orientation and site of integration are used to construct consolidated bacterial chromosome maps. The data below show the order of gene transfer for five strains.
Hfr Strain Order of Gene Transfer (First → Last)
Hfr A oriT–thr–leu–azi–ton–pro–lac–ade
Hfr B oriT–mtl–xyl–mal–str–his
Hfr C oriT–ile–met–thi–thr–leu–azi–ton
Hfr D oriT–his–trp–gal–ade–lac–pro–ton
Hfr E oriT–thi–met–ile–mtl–xyl–mal–str
Identify the overlaps between Hfr strains. Identify the orientations of integrated F factors relative to one another.
Problem 16b
Hfr strains that differ in integrated F factor orientation and site of integration are used to construct consolidated bacterial chromosome maps. The data below show the order of gene transfer for five strains.
Hfr Strain Order of Gene Transfer (First → Last)
Hfr A oriT–thr–leu–azi–ton–pro–lac–ade
Hfr B oriT–mtl–xyl–mal–str–his
Hfr C oriT–ile–met–thi–thr–leu–azi–ton
Hfr D oriT–his–trp–gal–ade–lac–pro–ton
Hfr E oriT–thi–met–ile–mtl–xyl–mal–str
Draw a consolidated map of the bacterial chromosome. (Hint: Begin by placing the insertion site for Hfr A at the 2 o'clock position and arranging the genes thr–leu–azi- . . . in clockwise order.)
Problem 17
Five Hfr strains from the same bacterial species are analyzed for their ability to transfer genes to F⁻ recipient bacteria. The data shown below list the origin of transfer (oriT) for each strain and give the order of genes, with the first gene on the left and the last gene on the right. Use the data to construct a circular map of the bacterium.
Hfr Strain Genes Transferred
Hfr 1 oriT met ala lac gal
Hfr 2 oriT met leu thr azi
Hfr 3 oriT gal pro trp azi
Hfr 4 oriT leu met ala lac
Hfr 5 oriT trp azi thr leu met
Problem 18a
An interrupted mating study is carried out on Hfr strains 1, 2, and 3 identified in Problem 17. After conjugation is established, a small sample of the mixture is collected every minute for 20 minutes to determine the distance between genes on the chromosome. Results for each of the three Hfr strains are shown below. The total duration of conjugation (in minutes) is given for each transferred gene.
Hfr strain 1 oriT met ala lac gal
Duration (min) 0 2 8 13 17
Hfr strain 2 oriT met leu thr azi
Duration (min) 0 2 7 10 17
Hfr strain 3 oriT gal pro trp azi
Duration (min) 0 3 8 14 19
For each Hfr strain, draw a time-of-entry profile.
Problem 18b
An interrupted mating study is carried out on Hfr strains 1, 2, and 3 identified in Problem 17. After conjugation is established, a small sample of the mixture is collected every minute for 20 minutes to determine the distance between genes on the chromosome. Results for each of the three Hfr strains are shown below. The total duration of conjugation (in minutes) is given for each transferred gene.
Hfr strain 1 oriT met ala lac gal
Duration (min) 0 2 8 13 17
Hfr strain 2 oriT met leu thr azi
Duration (min) 0 2 7 10 17
Hfr strain 3 oriT gal pro trp azi
Duration (min) 0 3 8 14 19
Using the chromosome map you prepared in answer to Problem 17, determine the distance in minutes between each gene on the map.
Problem 18c
An interrupted mating study is carried out on Hfr strains 1, 2, and 3 identified in Problem 17. After conjugation is established, a small sample of the mixture is collected every minute for 20 minutes to determine the distance between genes on the chromosome. Results for each of the three Hfr strains are shown below. The total duration of conjugation (in minutes) is given for each transferred gene.
Hfr strain 1 oriT met ala lac gal
Duration (min) 0 2 8 13 17
Hfr strain 2 oriT met leu thr azi
Duration (min) 0 2 7 10 17
Hfr strain 3 oriT gal pro trp azi
Duration (min) 0 3 8 14 19
Explain why azi is the last gene of strain 2 to transfer in the 20 minutes of conjugation time. How many minutes of conjugation time would be needed to allow the next gene on the map to transfer from Hfr strain 2?
Problem 18d
An interrupted mating study is carried out on Hfr strains 1, 2, and 3 identified in Problem 17. After conjugation is established, a small sample of the mixture is collected every minute for 20 minutes to determine the distance between genes on the chromosome. Results for each of the three Hfr strains are shown below. The total duration of conjugation (in minutes) is given for each transferred gene.
Hfr strain 1 oriT met ala lac gal
Duration (min) 0 2 8 13 17
Hfr strain 2 oriT met leu thr azi
Duration (min) 0 2 7 10 17
Hfr strain 3 oriT gal pro trp azi
Duration (min) 0 3 8 14 19
Write out the interrupted mating results you would expect after 20 minutes of conjugation for Hfr strains 4 and 5. Use the format shown at the beginning of this problem.
Problem 18e
An interrupted mating study is carried out on Hfr strains 1, 2, and 3 identified in Problem 17. After conjugation is established, a small sample of the mixture is collected every minute for 20 minutes to determine the distance between genes on the chromosome. Results for each of the three Hfr strains are shown below. The total duration of conjugation (in minutes) is given for each transferred gene.
Hfr strain 1 oriT met ala lac gal
Duration (min) 0 2 8 13 17
Hfr strain 2 oriT met leu thr azi
Duration (min) 0 2 7 10 17
Hfr strain 3 oriT gal pro trp azi
Duration (min) 0 3 8 14 19
In minutes, what is the total length of the chromosome in the donor species?
Problem 19
An Hfr strain with the genotype cys⁺ lue⁺ met⁺ strˢ is mated with an F- strain carrying the genotype cys⁻ lue⁻ met⁻ strᴿ. In an interrupted mating experiment, small samples of the conjugating bacteria are withdrawn every 3 minutes for 30 minutes. The withdrawn cells are shaken vigorously to stop conjugation and then placed on three different selection media, composed as follows:
Medium 1: Minimal medium plus leucine, methionine, and streptomycin
Medium 2: Minimal medium plus cysteine, methionine, and streptomycin
Medium 3: Minimal medium plus cysteine, leucine, and streptomycin
The following table shows the number of colonies growing on each selection medium. The sampling time indicates how many minutes have passed since conjugation began.
Sampling Time (minutes) Number of Colonies
Plate 1 Plate 2 Plate 3
3 0 0 0
6 0 0 0
9 0 62 0
12 0 87 0
15 51 124 0
18 79 210 62
21 109 250 85
24 144 250 111
27 152 250 122
30 152 250 122
Suppose a fourth selection medium containing leucine and streptomycin is prepared. At what sampling time do you expect the first-growing colonies to appear? Explain your reasoning.
Problem 19a
An Hfr strain with the genotype cys⁺ lue⁺ met⁺ strS is mated with an F- strain carrying the genotype cys⁻ lue⁻ met⁻ strᴿ. In an interrupted mating experiment, small samples of the conjugating bacteria are withdrawn every 3 minutes for 30 minutes. The withdrawn cells are shaken vigorously to stop conjugation and then placed on three different selection media, composed as follows:
Medium 1: Minimal medium plus leucine, methionine, and streptomycin
Medium 2: Minimal medium plus cysteine, methionine, and streptomycin
Medium 3: Minimal medium plus cysteine, leucine, and streptomycin
What donor gene is the selected marker in each medium?
Problem 19b
An Hfr strain with the genotype cys⁺ lue⁺ met⁺ strS is mated with an F- strain carrying the genotype cys⁻ lue⁻ met⁻ strᴿ. In an interrupted mating experiment, small samples of the conjugating bacteria are withdrawn every 3 minutes for 30 minutes. The withdrawn cells are shaken vigorously to stop conjugation and then placed on three different selection media, composed as follows:
Medium 1: Minimal medium plus leucine, methionine, and streptomycin
Medium 2: Minimal medium plus cysteine, methionine, and streptomycin
Medium 3: Minimal medium plus cysteine, leucine, and streptomycin
List all possible bacterial genotypes growing on each medium.
Problem 19c
An Hfr strain with the genotype cys⁺ lue⁺ met⁺ strS is mated with an F- strain carrying the genotype cys⁻ lue⁻ met⁻ strᴿ. In an interrupted mating experiment, small samples of the conjugating bacteria are withdrawn every 3 minutes for 30 minutes. The withdrawn cells are shaken vigorously to stop conjugation and then placed on three different selection media, composed as follows:
Medium 1: Minimal medium plus leucine, methionine, and streptomycin
Medium 2: Minimal medium plus cysteine, methionine, and streptomycin
Medium 3: Minimal medium plus cysteine, leucine, and streptomycin
What is the purpose of adding streptomycin to each selection medium?
Problem 19d
An Hfr strain with the genotype cys⁺ lue⁺ met⁺ strˢ is mated with an F- strain carrying the genotype cys⁻ lue⁻ met⁻ strᴿ. In an interrupted mating experiment, small samples of the conjugating bacteria are withdrawn every 3 minutes for 30 minutes. The withdrawn cells are shaken vigorously to stop conjugation and then placed on three different selection media, composed as follows:
Medium 1: Minimal medium plus leucine, methionine, and streptomycin
Medium 2: Minimal medium plus cysteine, methionine, and streptomycin
Medium 3: Minimal medium plus cysteine, leucine, and streptomycin
The following table shows the number of colonies growing on each selection medium. The sampling time indicates how many minutes have passed since conjugation began.
Sampling Time (minutes) Number of Colonies
Plate 1 Plate 2 Plate 3
3 0 0 0
6 0 0 0
9 0 62 0
12 0 87 0
15 51 124 0
18 79 210 62
21 109 250 85
24 144 250 111
27 152 250 122
30 152 250 122
Determine the order of donor genes cys, leu, and met from the interrupted mating data.
Problem 20
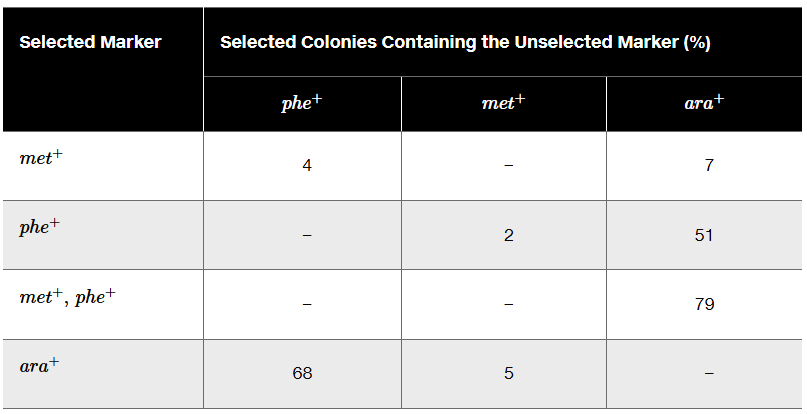
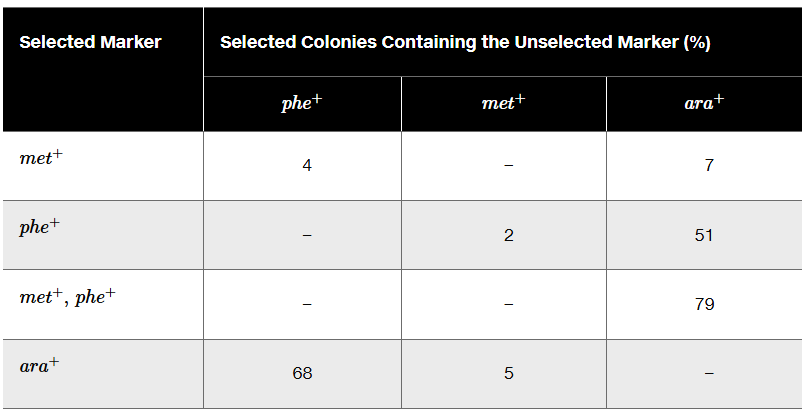
A triple-auxotrophic strain of E. coli having the genotype phe⁻ met⁻ ara⁻ is used as a recipient strain in a transduction experiment. The strain is unable to synthesize its own phenylalanine or methionine, and it carries a mutation that leaves it unable to utilize the sugar arabinose for growth. The recipient is crossed to a prototrophic strain with the genotype phe⁺ met⁺ ara⁺. The table below shows the selected marker and gives cotransduction frequencies for the unselected markers.
Use the cotransduction data to determine the order of these genes.
Problem 20a
A triple-auxotrophic strain of E. coli having the genotype phe⁻ met⁻ ara⁻ is used as a recipient strain in a transduction experiment. The strain is unable to synthesize its own phenylalanine or methionine, and it carries a mutation that leaves it unable to utilize the sugar arabinose for growth. The recipient is crossed to a prototrophic strain with the genotype phe⁺ met⁺ ara⁺. The table below shows the selected marker and gives cotransduction frequencies for the unselected markers.
Identify the compounds present in each of the selective media.
Problem 21a
Penicillin was first used in the 1940s to treat gonorrhea infections produced by the bacterium Neisseria gonorrhoeae. In 1984, according to the CDC, fewer than 1% of gonorrhea infections were caused by penicillin-resistant N. gonorrhoeae. By 1990, more than 10% of cases were penicillin-resistant, and a few years later the level of resistance was at greater than 95%. Almost every year the CDC issues new treatment guidelines for gonorrhea that identify the recommended antibiotic drugs and dosages.
Why is the CDC so active in making these recommendations?
Problem 21b
Penicillin was first used in the 1940s to treat gonorrhea infections produced by the bacterium Neisseria gonorrhoeae. In 1984, according to the CDC, fewer than 1% of gonorrhea infections were caused by penicillin-resistant N. gonorrhoeae. By 1990, more than 10% of cases were penicillin-resistant, and a few years later the level of resistance was at greater than 95%. Almost every year the CDC issues new treatment guidelines for gonorrhea that identify the recommended antibiotic drugs and dosages.
What are the short-term implications of these frequent changes for physicians and clinics that treat sexually transmitted diseases like gonorrhea and for individuals infected with gonorrhea?
Problem 21c
Penicillin was first used in the 1940s to treat gonorrhea infections produced by the bacterium Neisseria gonorrhoeae. In 1984, according to the CDC, fewer than 1% of gonorrhea infections were caused by penicillin-resistant N. gonorrhoeae. By 1990, more than 10% of cases were penicillin-resistant, and a few years later the level of resistance was at greater than 95%. Almost every year the CDC issues new treatment guidelines for gonorrhea that identify the recommended antibiotic drugs and dosages.
What are the long-term implications of these frequent changes in treatment recommendations for the patient population?
Problem 22a
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

How many genes are represented by these mutations?
Problem 22b
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

Identify the mutants of each gene.
Problem 22c
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

In each coinfection identified as a failure to complement (−) in the table, researchers see evidence of recombination producing wild-type growth. How do the researchers distinguish between wild-type growth resulting from complementation and wild-type growth that is due to recombination?
Problem 22d
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

A new mutation, designated 9, fails to complement mutants 1, 3, 5, 7, and 8. Wild-type recombinants form between mutant 9 and mutations 3, 5, and 8; however, no wild-type recombinants form between mutant 9 and mutations 1 and 7. What kind of mutation is mutant 9? Explain your reasoning.
Problem 22e
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

New mutation 10 fails to complement mutants 1, 4, 5, 6, 8, and 9. Mutant 10 forms wild-type recombinants with mutants 1, 5, and 6, but not with mutants 4 and 8. Mutant 9 and mutant 10 form wild-type recombinants. What kind of mutation is mutant 10? Explain your reasoning.