Back
BackProblem 22f
An attribute of growth behavior of eight bacteriophage mutants (1 to 8) is investigated in experiments that establish coinfection by pairs of mutants. The experiments determine whether the mutants complement one another (+) or fail to complement (-). These eight mutants are known to result from point mutation. The results of the complementation tests are shown below.

Gene-mapping information identifies mutations 2 and 3 as the flanking markers in this group of genes. Assuming these mutations are on opposite ends of the gene map, determine the order of mutations in the region of the chromosome.
Problem 23a
Synthesis of the amino acid histidine is a multistep anabolic pathway that uses the products of 13 genes (hisA to hisM) in E. coli. Two independently isolated his- E. coli mutants, designated his1⁻ and his2⁻ are studied in a conjugation experiment. A his⁺ F' donor strain that carries a copy of the hisJ gene on the plasmid is mated with a his1⁻ recipient strain in Experiment 1 and with a his2⁻ recipient in Experiment 2. The exconjugants are grown on plates lacking histidine. Growth is observed among the exconjugants of Experiment 2 but not among those of Experiment 1.
Why is growth observed in Experiment 2 but not in Experiment 1?
Problem 23b
Synthesis of the amino acid histidine is a multistep anabolic pathway that uses the products of 13 genes (hisA to hisM) in E. coli. Two independently isolated his- E. coli mutants, designated his1⁻ and his2⁻ are studied in a conjugation experiment. A his⁺ F' donor strain that carries a copy of the hisJ gene on the plasmid is mated with a his1⁻ recipient strain in Experiment 1 and with a his2⁻ recipient in Experiment 2. The exconjugants are grown on plates lacking histidine. Growth is observed among the exconjugants of Experiment 2 but not among those of Experiment 1.
What is the genotype of exconjugants in Experiment 2?
Problem 24a
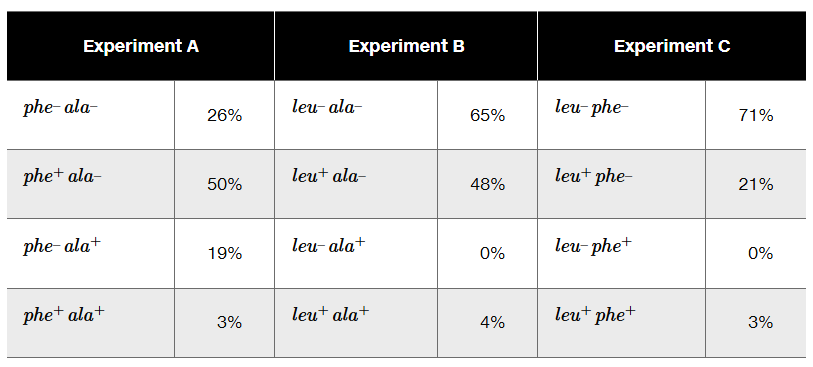
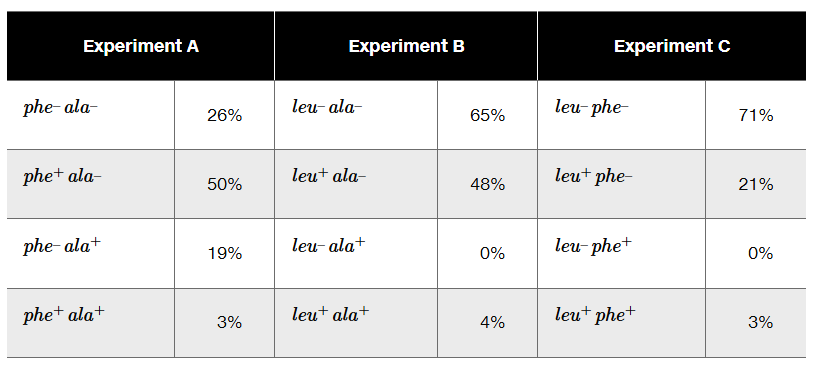
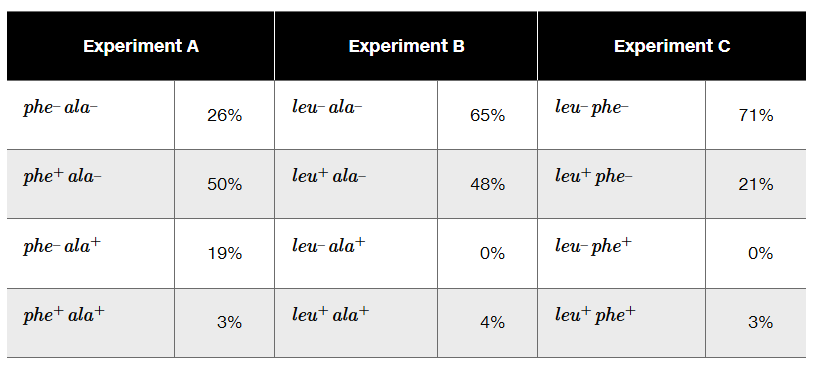
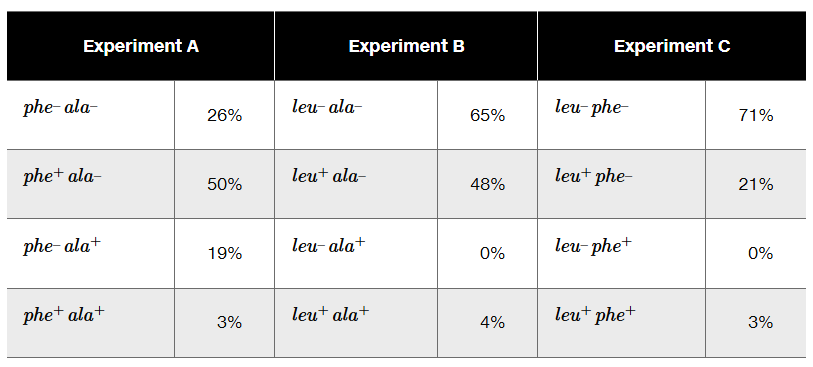
The phage P1 is used as a generalized transducing phage in an experiment combining a donor strain of E. coli of genotype leu⁺ phe⁺ ala⁺ and a recipient strain that is leu⁻ phe⁻ ala⁻. In separate experiments, transductants are selected for leu⁺ (Experiment A), for ala⁺ (Experiment B), and for phe⁺ (Experiment C). Following selection, transductant genotypes for the unselected markers are identified. The selection experiment results below show the frequency of each genotype.
What compound or compounds are added to the minimal medium to select for transductants in Experiments A, B, and C?
Problem 24b
The phage P1 is used as a generalized transducing phage in an experiment combining a donor strain of E. coli of genotype leu⁺ phe⁺ ala⁺ and a recipient strain that is leu⁻ phe⁻ ala⁻. In separate experiments, transductants are selected for leu⁺ (Experiment A), for ala⁺ (Experiment B), and for phe⁺ (Experiment C). Following selection, transductant genotypes for the unselected markers are identified. The selection experiment results below show the frequency of each genotype.
Determine the order of genes on the donor chromosome.
Problem 24c
The phage P1 is used as a generalized transducing phage in an experiment combining a donor strain of E. coli of genotype leu⁺ phe⁺ ala⁺ and a recipient strain that is leu⁻ phe⁻ ala⁻. In separate experiments, transductants are selected for leu⁺ (Experiment A), for ala⁺ (Experiment B), and for phe⁺ (Experiment C). Following selection, transductant genotypes for the unselected markers are identified. The selection experiment results below show the frequency of each genotype.
Diagram the crossover events that form each of the transductants in Experiment A.
Problem 24d
The phage P1 is used as a generalized transducing phage in an experiment combining a donor strain of E. coli of genotype leu⁺ phe⁺ ala⁺ and a recipient strain that is leu⁻ phe⁻ ala⁻. In separate experiments, transductants are selected for leu⁺ (Experiment A), for ala⁺ (Experiment B), and for phe⁺ (Experiment C). Following selection, transductant genotypes for the unselected markers are identified. The selection experiment results below show the frequency of each genotype.
In Experiment B, why are there no transductants with the genotype leu⁻ ala⁺?
Problem 25
Problem 25a
Define the term genetic complementation.
Describe how the term applies to an experiment in which two lysis-defective bacteriophages are able to coinfect a bacterial cell and produce lysis.
Problem 25b
Define the term genetic complementation.
Give another example of genetic complementation and describe how genetic complementation works in that case.
Problem 26
Devise an experiment to identify bacteria that are auxotrophic and unable to produce two amino acids, lysine (lys) and valine (val). The auxotrophic bacteria are in a pool of bacteria in which all the other bacteria are prototrophic. The genotype of the auxotrophs is lys⁻ val⁻. Describe each step in the experiment, identify the constituents in any growth medium or growth plates you propose, and identify the results that will conclusively identify bacteria that are lys⁻ val⁻.
Problem 27
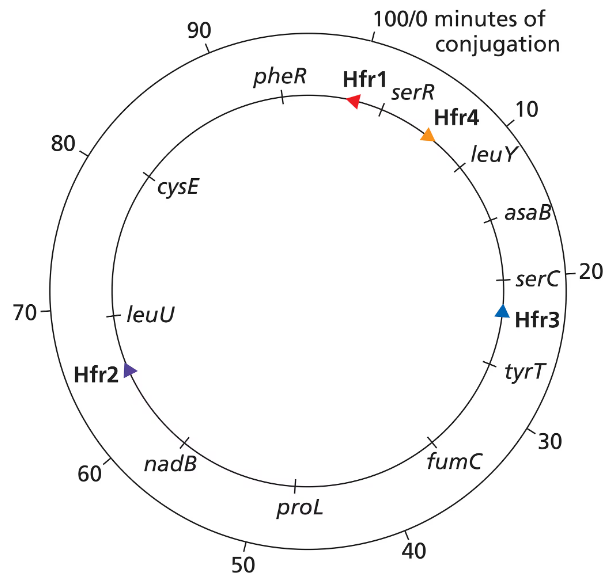
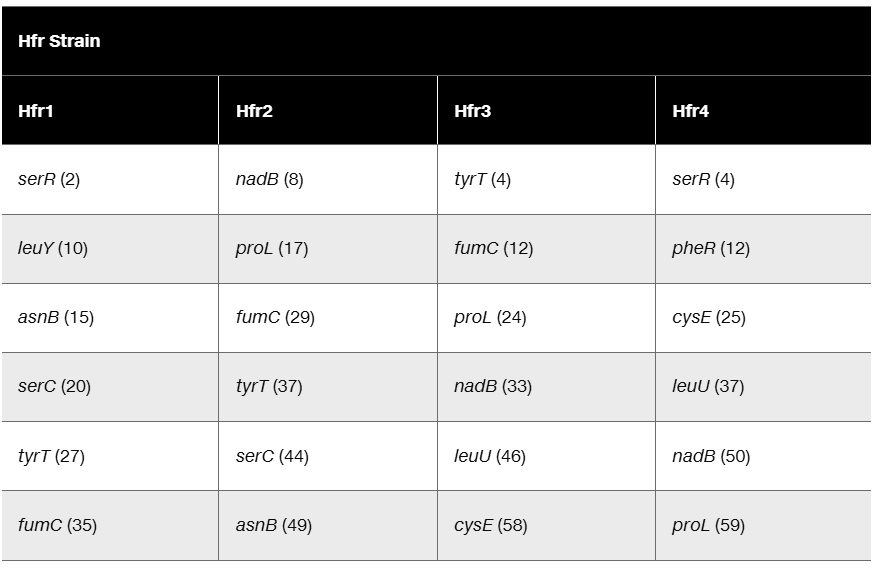
Look closely at the consolidated Hfr map and the data used to build the map on page 261. Suppose a fifth Hfr strain had the F factor inserted exactly halfway between cysE and leuU and had an orientation that was the same as that of Hfr 1. List the order of gene transfer for the first six genes transferred by this Hfr and the number of minutes of conjugation at which each gene is expected to be seen.
Problem 28
Fifty bacterial colonies are on a complete-medium growth plate. The colonies are replica plated to a minimal-medium plate, and 46 colonies grow. What can you say about the bacteria from the four colonies that do not grow? Design an experiment and describe the methods you would use to determine if any of these four colonies are leu⁻, arg⁻, or val⁻.