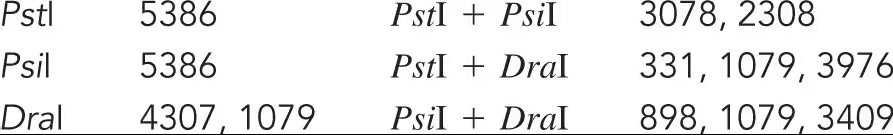Back
BackProblem 11
Injection of double-stranded RNA can lead to gene silencing by degradation of RNA molecules complementary to either strand of the dsRNA. Could RNAi be used in gene therapy for a defect caused by a recessive allele? A dominant allele? If so, what might be the major obstacle to using RNAi as a therapeutic agent?
Problem E.11a
In an inheritance case, a man has died leaving his estate to be divided equally between 'his wife and his offspring.' His wife (M) has an adult daughter (D), and they argue that they should split the estate equally. As a young couple, however, the man and his wife had a son that they gave up for adoption. Two men have appeared, each claiming to be the son of the couple and therefore entitled to a one-third share of the estate. The accompanying illustration shows the results of DNA analysis for five genes for the mother (M), her daughter (D), and the two claimants (S1 and S2). How many nonmaternal DNA bands are shared by D and S1? By D and S2?
Problem E.11b
In an inheritance case, a man has died leaving his estate to be divided equally between 'his wife and his offspring.' His wife (M) has an adult daughter (D), and they argue that they should split the estate equally. As a young couple, however, the man and his wife had a son that they gave up for adoption. Two men have appeared, each claiming to be the son of the couple and therefore entitled to a one-third share of the estate. The accompanying illustration shows the results of DNA analysis for five genes for the mother (M), her daughter (D), and the two claimants (S1 and S2). Do the DNA results suggest that either man is likely to be the son of the man and his wife? Explain.
Problem 12
Compare and contrast methods for making transgenic plants and transgenic Drosophila.
Problem E.12a
Three independently assorting STR markers (A, B, and C) are used to assess the paternity of a colt recently born to a quarter horse mare. Blood samples are drawn from the mare, her colt, and three possible male sires (S₁, S₂, and S₃). DNA at each marker locus is amplified by PCR, and a DNA electrophoresis gel is run for each marker. Amplified DNA bands are visualized in each gel by ethidium bromide staining. Gel results are shown below for each marker. Evaluate the data and determine if any of the potential sires can be excluded. Explain the basis of exclusion, if any, in each case.
Problem E.12b
Three independently assorting STR markers (A, B, and C) are used to assess the paternity of a colt recently born to a quarter horse mare. Blood samples are drawn from the mare, her colt, and three possible male sires (S₁, S₂, and S₃). DNA at each marker locus is amplified by PCR, and a DNA electrophoresis gel is run for each marker. Amplified DNA bands are visualized in each gel by ethidium bromide staining. Gel results are shown below for each marker. Calculate the PI and CPI based on these STR markers, using the following population frequencies: A₁₂ = 0.12, A₁₀ = 0.18; B₁₈ = 0.08, B₁₂ = 0.17; C₁₆ = 0.11, C₁₄ = 0.20.
- It is often desirable to insert cDNAs into a cloning vector in such a way that all the cDNA clones will have the same orientation with respect to the sequences of the plasmid. This is referred to as directional cloning. Outline how you would directionally clone a cDNA library in the plasmid vector pUC18.
Problem 13
- A major advance in the 1980s was the development of technology to synthesize short oligonucleotides. This work both facilitated DNA sequencing and led to the advent of the development of PCR. Recently, rapid advances have occurred in the technology to chemically synthesize DNA, and sequences up to 10 kb are now readily produced. As this process becomes more economical, how will it affect the gene-cloning approaches outlined in this chapter? In other words, what types of techniques does this new technology have potential to supplant, and what techniques will not be affected by it?
Problem 14
Problem 15a
The bacteriophage lambda genome can exist in either a linear form or a circular form.
How many fragments will be formed by restriction enzyme digestion with XhoI alone, with XbaI alone, and with both XhoI and XbaI in the linear and circular forms of the lambda genome?
Problem 15b
The bacteriophage lambda genome can exist in either a linear form or a circular form.
Diagram the resulting fragments as they would appear on an agarose gel after electrophoresis.
Problem 16
The restriction enzymes XhoI and SalI cut their specific sequences as shown below:
Can the sticky ends created by XhoI and SalI sites be ligated? If yes, can the resulting sequences be cleaved by either XhoI or SalI?
Problem 17
The bacteriophage ϕX174 has a single-stranded DNA genome of 5386 bases. During DNA replication, double-stranded forms of the genome are generated. In an effort to create a restriction map of ϕX174, you digest the z-stranded form of the genome with several restriction enzymes and obtain the following results. Draw a map of the ϕX174 genome.
Problem 18
To further analyze the CRABS CLAW gene, you create a map of the genomic clone. The 11-kb EcoRI fragment is ligated into the EcoRI site of the MCS of the vector shown in Problem 18. You digest the double-stranded form of the genome with several restriction enzymes and obtain the following results. Draw, as far as possible, a map of the genomic clone of CRABS CLAW.
What restriction digest would help resolve any ambiguity in the map?
Problem 19a
You have isolated a genomic clone with an EcoRI fragment of 11 kb that encompasses the CRABS CLAW gene. You digest the genomic clone with HindIII and note that the 11-kb EcoRI fragment is split into three fragments of 9 kb, 1.5 kb, and 0.5 kb.
Does this tell you anything about where the CRABS CLAW gene is located within the 11-kb genomic clone?
Problem 19b
You have isolated a genomic clone with an EcoRI fragment of 11 kb that encompasses the CRABS CLAW gene (see Problem 18). You digest the genomic clone with HindIII and note that the 11-kb EcoRI fragment is split into three fragments of 9 kb, 1.5 kb, and 0.5 kb.
Restriction enzyme sites within a cDNA clone are often also found in the genomic sequence. Can you think of a reason why occasionally this is not the case? What about the converse: Are restriction enzyme sites in a genomic clone always in a cDNA clone of the same gene?
Problem 20
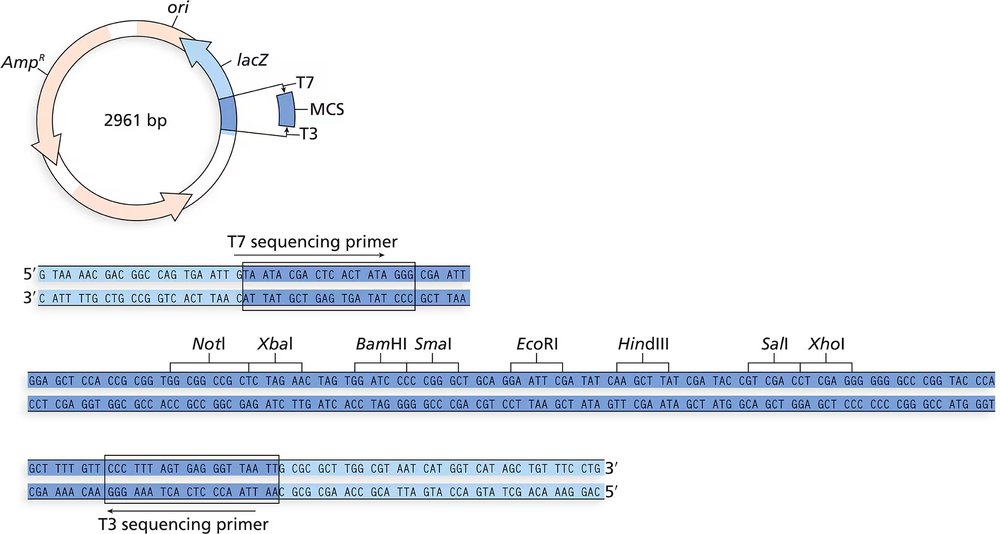
You have identified a 0.80-kb cDNA clone that contains the entire coding sequence of the Arabidopsis gene CRABS CLAW. In the construction of the cDNA library, linkers with EcoRI sites were added to each end of the cDNA, and the cDNA was inserted into the EcoRI site of the MCS of the vector shown in the accompanying figure. You perform digests on the CRABS CLAW cDNA clone with restriction enzymes and obtain the following results. Can you determine the orientation of the cDNA clone with respect to the restriction enzyme sites in the vector? The restriction enzyme sites listed in the dark blue region are found only in the MCS of the vector.
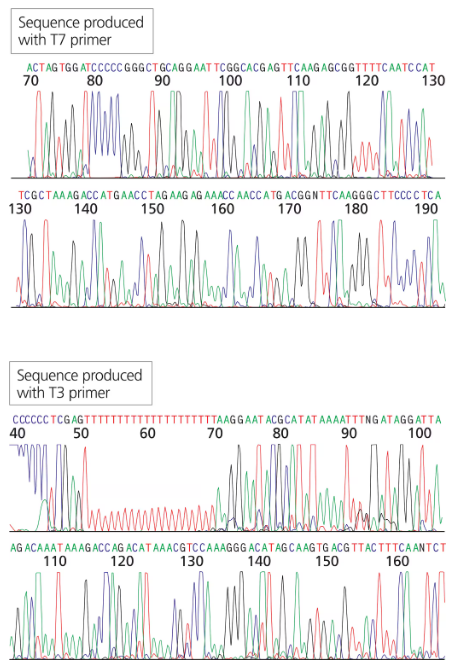
Problem 21b
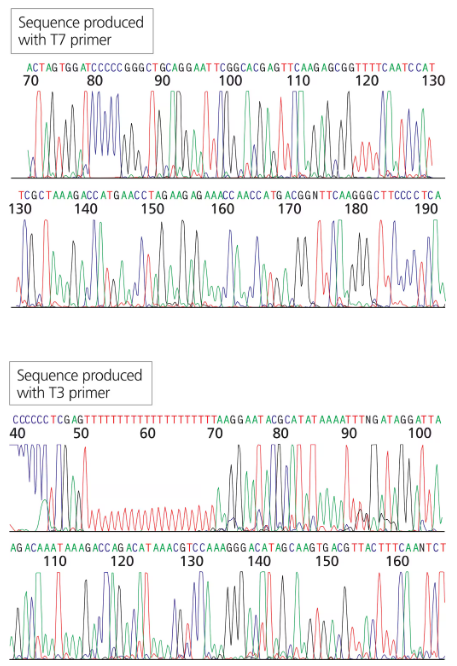
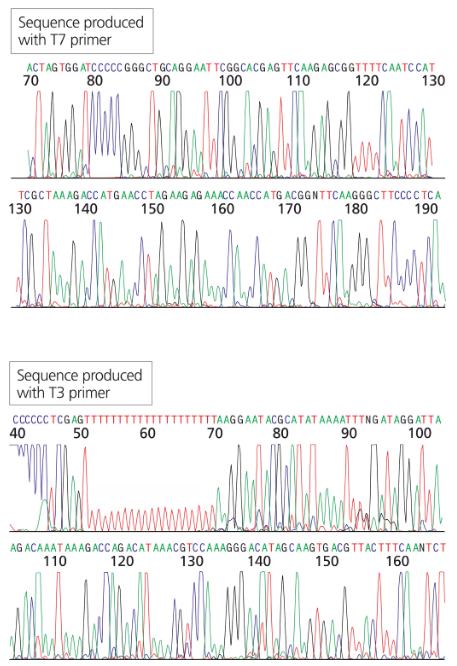
You have isolated another cDNA clone of the CRABS CLAW gene from a cDNA library. The cDNA was directionally cloned using the EcoRI and XhoI sites. You sequence the recombinant plasmid using primers complementary to the T7 and T3 promoter sites flanking the MCS. The first 30 to 60 bases of sequence are usually discarded since they tend to contain errors.
Will the long stretch of T residues in the T3 sequence exist in the genomic sequence of the gene?
Problem 21c
You have isolated another cDNA clone of the CRABS CLAW gene from a cDNA library.. The cDNA was directionally cloned using the EcoRI and XhoI sites. You sequence the recombinant plasmid using primers complementary to the T7 and T3 promoter sites flanking the MCS. The first 30 to 60 bases of sequence are usually discarded since they tend to contain errors.
Can you identify which sequence portions are derived from the vector (specifically the MCS) and which are derived from the cDNA clone?
Problem 21d
You have isolated another cDNA clone of the CRABS CLAW gene from a cDNA library. The cDNA was directionally cloned using the EcoRI and XhoI sites. You sequence the recombinant plasmid using primers complementary to the T7 and T3 promoter sites flanking the MCS. The first 30 to 60 bases of sequence are usually discarded since they tend to contain errors.
Can you identify the start of the coding region in the end of the gene? What does the sequence preceding the start codon represent?
Problem 22a
You have identified five genes in S. cerevisiae that are induced when the yeast are grown in a high-salt (NaCl) medium. To study the potential roles of these genes in acclimation to growth in high-salt conditions, you wish to examine the phenotypes of loss- and gain-of-function alleles of each. How will you do this?
Problem 22b
You have identified five genes in S. cerevisiae that are induced when the yeast are grown in a high-salt (NaCl) medium. To study the potential roles of these genes in acclimation to growth in high-salt conditions, you wish to examine the phenotypes of loss- and gain-of-function alleles of each. How would your answer differ if you were working with tomato plants instead of yeast?
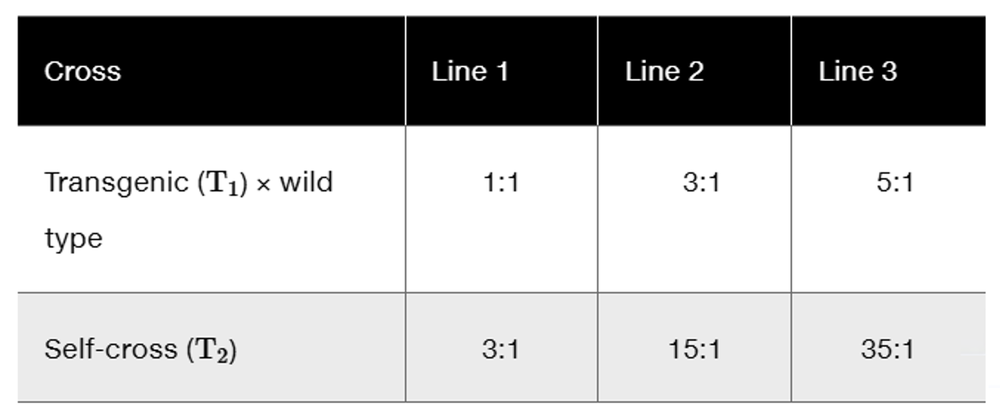
Problem 23
You have generated three transgenic lines of maize that are resistant to the European corn borer, a significant pest in many regions of the world. The transgenic lines (T₁ in the accompanying table) were created using Agrobacterium-mediated transformation with a T-DNA having two genes, the first being a gene conferring resistance to the corn borer and the second being a gene conferring resistance to a herbicide that you used as a selectable marker to obtain your transgenic plants. You crossed each of the lines to a wild-type maize plant and also generated a T2 population by self-fertilization of the T1 plant. The following segregation results were observed (herbicide resistant : herbicide sensitive):
Explain these segregation ratios.
Problem 24
Bacterial Pseudomonas species often possess plasmids encoding genes involved in the catabolism of organic compounds. You have discovered a strain that can metabolize crude oil and wish to identify the gene(s) responsible. Outline an experimental protocol to find the gene or genes required for crude oil metabolism.
Problem 25
Two complaints about some transgenic plants presently in commercial use are that (1) the Bt toxin gene is constitutively expressed in them, leading to fears that selection pressures will cause insects to evolve resistance to the toxin, and (2) a selectable marker gene—for example, conferring kanamycin resistance—remains in the plant, leading to concerns about increased antibiotic resistance in organisms in the wild. How would you generate transgenic plants that produce Bt only in response to being fed upon by insects and without the selectable marker?
Problem 26a
In Drosophila, loss-of-function Ultrabithorax mutations result in the posterior thoracic segments differentiating into body parts with an identity normally found in the anterior thoracic segments. When the Ultrabithorax gene was cloned, it was shown to encode a transcription factor and to be expressed only in the posterior region of the thorax. Thus, Ultrabithorax acts to specify the identity of the posterior thoracic segments. Similar genes were soon discovered in other animals, including mice and humans. You have found that mice possess two closely related genes, Hoxa7 and Hoxb4, which are orthologs of Ultrabithorax. You wish to know whether the two mouse genes act to specify the identity of body segments in mice.
How will you determine where and when the mouse genes are expressed?
Problem 26b
In Drosophila, loss-of-function Ultrabithorax mutations result in the posterior thoracic segments differentiating into body parts with an identity normally found in the anterior thoracic segments. When the Ultrabithorax gene was cloned, it was shown to encode a transcription factor and to be expressed only in the posterior region of the thorax. Thus, Ultrabithorax acts to specify the identity of the posterior thoracic segments. Similar genes were soon discovered in other animals, including mice and humans. You have found that mice possess two closely related genes, Hoxa7 and Hoxb4, which are orthologs of Ultrabithorax. You wish to know whether the two mouse genes act to specify the identity of body segments in mice.
How will you create loss-of-function alleles of the mouse genes?
Problem 26c
In Drosophila, loss-of-function Ultrabithorax mutations result in the posterior thoracic segments differentiating into body parts with an identity normally found in the anterior thoracic segments. When the Ultrabithorax gene was cloned, it was shown to encode a transcription factor and to be expressed only in the posterior region of the thorax. Thus, Ultrabithorax acts to specify the identity of the posterior thoracic segments. Similar genes were soon discovered in other animals, including mice and humans. You have found that mice possess two closely related genes, Hoxa7 and Hoxb4, which are orthologs of Ultrabithorax. You wish to know whether the two mouse genes act to specify the identity of body segments in mice.
How will you determine whether the mouse genes have redundant functions?
Problem 27a
You have identified an enhancer trap line generated by P element transposition in Drosophila in which the marker gene from the enhancer trap is specifically expressed in the wing imaginal disc.
How can you identify the gene adjacent to the insertion site of the enhancer trap?
Problem 27b
You have identified an enhancer trap line generated by P element transposition in Drosophila in which the marker gene from the enhancer trap is specifically expressed in the wing imaginal disc.
How would you show that the expression pattern of the enhancer trap line reflects the endogenous gene expression pattern of the adjacent gene?
Problem 28
The highlighted sequence shown below is the one originally used to produce the B chain of human insulin in E. coli. The sequence of the human gene encoding the B chain of insulin was later determined from a cDNA isolated from a human pancreatic cDNA library and is also shown below, without highlighting. Explain the differences between the two sequences.
ATGTTCGTCAATCAGCACCTTTGTGGTTCTCACCTCGTTGAAGCTTTGTACCTTGTTTGCGGTGAACGTGGTTTCTTCTACACTCCTAAGACTTAA
GCCTTTGTGAACCAACACCTGTGCGGCTCACACCTGGTGGAAGCTCTCTACCTAGTGTGCGGGGAACGAGGCTTCTTCTACACACCCAAGACCCGC