You have identified a recessive mutation that alters bristle patterning in Drosophila and have used recombinant DNA technology to identify a genomic clone that you believe harbors the gene. How would you demonstrate that your gene is on the genomic clone?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 14 - Analysis of Gene Function via Forward Genetics and Reverse Genetics
Ch. 14 - Analysis of Gene Function via Forward Genetics and Reverse Genetics Problem 13b
Problem 13bThe CBF genes of Arabidopsis are induced by exposure of the plants to low temperature. Can you design a method that would reveal these changes in gene expression in a way that a farmer could recognize them by observing plants growing in the field?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Gene Expression

Phenotypic Changes
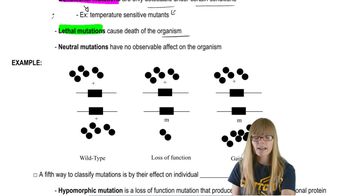
Field-Based Observation Techniques
Genetic counseling has not been discussed in this chapter, but it is a service provided by trained professional counselors who also have detailed knowledge of medical genetics, as described in Application Chapter A. Genetic counselors provide details about gene mutations and have knowledge of most of the details of diseases associated with genetic abnormalities. With regard to genetic testing to identify one's personal risk of cancer, what are the three or four topics you think are most important to be able to discuss with a genetic counselor?
The CBF genes of Arabidopsis are induced by exposure of the plants to low temperature. How would you examine the temporal and spatial patterns of expression after induction by low temperature?
When the S. cerevisiae genome was sequenced and surveyed for possible genes, only about 40% of those genes had been previously identified in forward genetic screens. This left about 60% of predicted genes with no known function, leading some to dub the genes fun (function unknown) genes. As an approach to understanding the function of a certain fun gene, you wish to create a loss-of-function allele. How will you accomplish this?
When the S. cerevisiae genome was sequenced and surveyed for possible genes, only about 40% of those genes had been previously identified in forward genetic screens. This left about 60% of predicted genes with no known function, leading some to dub the genes fun (function unknown) genes. You wish to know the physical location of the encoded protein product. How will you obtain such information?
Translational fusions between a protein of interest and a reporter protein are used to determine the subcellular location of proteins in vivo. However, fusion to a reporter protein sometimes renders the protein of interest nonfunctional because the addition of the reporter protein interferes with proper protein folding, enzymatic activity, or protein–protein interactions. You have constructed a fusion between your protein of interest and a reporter gene. How will you show that the fusion protein retains its normal biological function?