Back
BackProblem 24
Describe the role of two forms of RNA editing that lead to changes in the size and sequence of pre-mRNAs. Briefly describe several examples of each form of editing, including their impact on respective protein products.
Problem 25
Substitution RNA editing is known to involve either C-to-U or A-to-I conversions. What common chemical event accounts for each?
Problem 26a
It has been suggested that the present-day triplet genetic code evolved from a doublet code when there were fewer amino acids available for primitive protein synthesis.
Can you find any support for the doublet code notion in the existing coding dictionary?
Problem 26b
It has been suggested that the present-day triplet genetic code evolved from a doublet code when there were fewer amino acids available for primitive protein synthesis.
The amino acids Ala, Val, Gly, Asp, and Glu are all early members of biosynthetic pathways and are more evolutionarily conserved than other amino acids. They therefore probably represent 'early' amino acids. Of what significance is this information in terms of the evolution of the genetic code? Also, which base, of the first two within a coding triplet, would likely have been the more significant in originally specifying these amino acids?
Problem 26c
It has been suggested that the present-day triplet genetic code evolved from a doublet code when there were fewer amino acids available for primitive protein synthesis.
As determined by comparisons of ancient and recently evolved proteins, cysteine, tyrosine, and phenylalanine appear to be late-arriving amino acids. In addition, they are considered to have been absent in the abiotic Earth. All three of these amino acids have only two codons each, while many others, earlier in origin, have more. Is this mere coincidence, or might there be some underlying explanation?
Problem 27
An early proposal by George Gamow in 1954 regarding the genetic code considered the possibility that DNA served directly as the template for polypeptide synthesis. In eukaryotes, what difficulties would such a system pose? What observations and theoretical considerations argue against such a proposal?
Problem 28
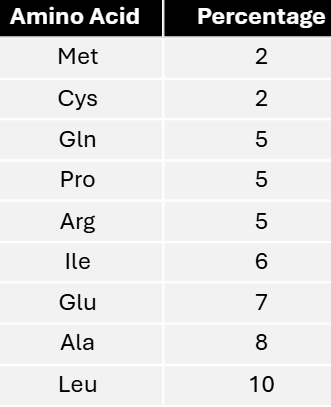
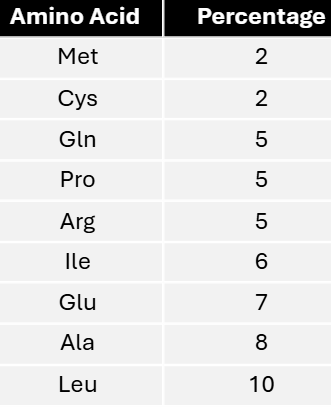
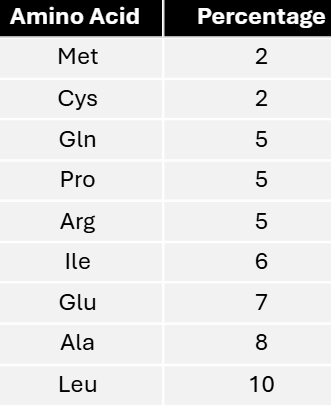
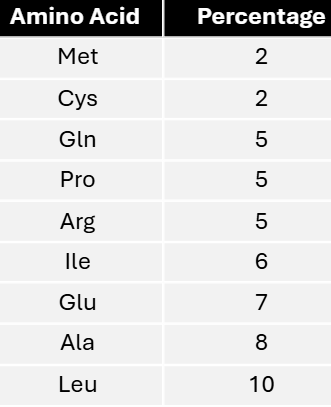
In a mixed copolymer experiment, messages were created with either 4/5C:1/5A or 4/5A:1/5C. These messages yielded proteins with the following amino acid compositions.

Using these data, predict the most specific coding composition for each amino acid.
Problem 29a
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-His _
Use this information to answer the following questions:
Using the genetic coding dictionary, predict the type of mutation that led to each altered protein.
Problem 29b
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-Hi
___________________________________________________
Use this information to answer the following questions:
For each mutant protein, determine the specific ribonucleotide change that led to its synthesis.
Problem 29c
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
__________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met -Cys-Ile-Val-Val-Val-Gln-His
______________________________________________
Use this information to answer the following questions:
The wild-type RNA consists of nine triplets. What is the role of the ninth triplet?
Problem 29d
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-Hi
___________________________________________________
Use this information to answer the following questions:
Of the first eight wild-type triplets, which, if any, can you determine specifically from an analysis of the mutant proteins? In each case, explain why or why not.
Problem 29e
Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met -Cys-Ile-Val-Val-Val-Gln-His
___________________________________________________
Use this information to answer the following questions:
Another mutation (mutant 4) is isolated. Its amino acid sequence is unchanged from the wild type, but the mutant cells produce abnormally low amounts of the wild-type proteins. As specifically as you can, predict where this mutation exists in the gene.
Problem 30a
The genetic code is degenerate. Amino acids are encoded by either 1, 2, 3, 4, or 6 triplet codons. An interesting question is whether the number of triplet codes for a given amino acid is in any way correlated with the frequency with which that amino acid appears in proteins. That is, is the genetic code optimized for its intended use? Some approximations of the frequency of appearance of nine amino acids in proteins in E. coli are given in the following:
Determine how many triplets encode each amino acid.
Problem 30b
The genetic code is degenerate. Amino acids are encoded by either 1, 2, 3, 4, or 6 triplet codons. An interesting question is whether the number of triplet codes for a given amino acid is in any way correlated with the frequency with which that amino acid appears in proteins. That is, is the genetic code optimized for its intended use? Some approximations of the frequency of appearance of nine amino acids in proteins in E. coli are given in the following:
Devise a way to graphically compare the two sets of information (data).
Problem 30c
The genetic code is degenerate. Amino acids are encoded by either 1, 2, 3, 4, or 6 triplet codons. An interesting question is whether the number of triplet codes for a given amino acid is in any way correlated with the frequency with which that amino acid appears in proteins. That is, is the genetic code optimized for its intended use? Some approximations of the frequency of appearance of nine amino acids in proteins in E. coli are given in the following:
Analyze your data to determine what, if any, correlations can be drawn between the relative frequency of amino acids making up proteins and the number of codons for each. Write a paragraph that states your specific and general conclusions.
Problem 30d
The genetic code is degenerate. Amino acids are encoded by either 1, 2, 3, 4, or 6 triplet codons. An interesting question is whether the number of triplet codes for a given amino acid is in any way correlated with the frequency with which that amino acid appears in proteins. That is, is the genetic code optimized for its intended use? Some approximations of the frequency of appearance of nine amino acids in proteins in E. coli are given in the following:
How would you proceed with your analysis if you wanted to pursue this problem further?
Problem 31a
M. Klemke et al. (2001) discovered an interesting coding phenomenon in which an exon within a neurologic hormone receptor gene in mammals appears to produce two different protein entities (and ALEX). The following is the DNA sequence of the exon's end derived from a rat.
5'-gtcccaaccatgcccaccgatcttccgcctgcttctgaagATGCGGGCCCAG
The lowercase letters represent the initial coding portion for the protein, and the uppercase letters indicate the portion where the ALEX entity is initiated. (For simplicity, and to correspond with the RNA coding dictionary, it is customary to represent the coding (non-template) strand of the DNA segment.)
Convert the coding DNA sequence to the coding RNA sequence.
Problem 31b
M. Klemke et al. (2001) discovered an interesting coding phenomenon in which an exon within a neurologic hormone receptor gene in mammals appears to produce two different protein entities (and ALEX). The following is the DNA sequence of the exon's end derived from a rat.
5'-gtcccaaccatgcccaccgatcttccgcctgcttctgaagATGCGGGCCCAG
The lowercase letters represent the initial coding portion for the protein, and the uppercase letters indicate the portion where the ALEX entity is initiated. (For simplicity, and to correspond with the RNA coding dictionary, it is customary to represent the coding (non-template) strand of the DNA segment.)
Locate the initiator codon within the XLαs segment.
Problem 31c
M. Klemke et al. (2001) discovered an interesting coding phenomenon in which an exon within a neurologic hormone receptor gene in mammals appears to produce two different protein entities (and ALEX). The following is the DNA sequence of the exon's end derived from a rat.
5'-gtcccaaccatgcccaccgatcttccgcctgcttctgaagATGCGGGCCCAG
The lowercase letters represent the initial coding portion for the protein, and the uppercase letters indicate the portion where the ALEX entity is initiated. (For simplicity, and to correspond with the RNA coding dictionary, it is customary to represent the coding (non-template) strand of the DNA segment.)
Locate the initiator codon within the ALEX segment. Are the two initiator codons in frame?
Problem 31d
M. Klemke et al. (2001) discovered an interesting coding phenomenon in which an exon within a neurologic hormone receptor gene in mammals appears to produce two different protein entities (and ALEX). The following is the DNA sequence of the exon's end derived from a rat.
5'-gtcccaaccatgcccaccgatcttccgcctgcttctgaagATGCGGGCCCAG
The lowercase letters represent the initial coding portion for the protein, and the uppercase letters indicate the portion where the ALEX entity is initiated. (For simplicity, and to correspond with the RNA coding dictionary, it is customary to represent the coding (non-template) strand of the DNA segment.)
Provide the amino acid sequence for each coding sequence. In the region of overlap, are the two amino acid sequences the same?
Problem 31e
M. Klemke et al. (2001) discovered an interesting coding phenomenon in which an exon within a neurologic hormone receptor gene in mammals appears to produce two different protein entities (and ALEX). The following is the DNA sequence of the exon's end derived from a rat.
5'-gtcccaaccatgcccaccgatcttccgcctgcttctgaagATGCGGGCCCAG
The lowercase letters represent the initial coding portion for the protein, and the uppercase letters indicate the portion where the ALEX entity is initiated. (For simplicity, and to correspond with the RNA coding dictionary, it is customary to represent the coding (non-template) strand of the DNA segment.)
Are there any evolutionary advantages to having the same DNA sequence code for two protein products? Are there any disadvantages?
Problem 32b
Recent observations indicate that alternative splicing is a common way for eukaryotes to expand their repertoire of gene functions. Studies indicate that approximately 50 percent of human genes exhibit alternative splicing and approximately 15 percent of disease-causing mutations involve aberrant alternative splicing. Different tissues show remarkably different frequencies of alternative splicing, with the brain accounting for approximately 18 percent of such events [Xu et al. (2002). Nucl. Acids Res. 30:3754–3766].
Why might some tissues engage in more alternative splicing than others?
Problem 33a
Isoginkgetin is a cell-permeable chemical isolated from the Ginkgo biloba tree that binds to and inhibits snRNPs.
What types of problems would you anticipate in cells treated with isoginkgetin?
Problem 33b
Isoginkgetin is a cell-permeable chemical isolated from the Ginkgo biloba tree that binds to and inhibits snRNPs.
Would this be most problematic for E. coli cells, yeast cells, or human cells? Why?