Back
BackProblem 30d
Drosophila has a diploid chromosome number of 2n = 8, which includes one pair of sex chromosomes (XX in females and XY in males) and three pairs of autosomes. Consider a Drosophila male that has a copy of the A₁ allele on its X chromosome (the Y chromosome is the homolog) and is heterozygous for alleles B₁ and B₂, C₁ and C₂, and D₁ and D₂ of genes that are each on a different autosomal pair. In the diagrams requested below, indicate the alleles carried on each chromosome and sister chromatid. Assume that no crossover occurs between homologous chromosomes.
For the metaphase I alignment shown in (c), what gamete genotypes are produced at the end of meiosis?
Problem 30e
Drosophila has a diploid chromosome number of 2n = 8, which includes one pair of sex chromosomes (XX in females and XY in males) and three pairs of autosomes. Consider a Drosophila male that has a copy of the A₁ allele on its X chromosome (the Y chromosome is the homolog) and is heterozygous for alleles B₁ and B₂, C₁ and C₂, and D₁ and D₂ of genes that are each on a different autosomal pair. In the diagrams requested below, indicate the alleles carried on each chromosome and sister chromatid. Assume that no crossover occurs between homologous chromosomes.
How many different metaphase I chromosome alignments are possible in this male? How many genetically different gametes can this male produce? Explain your reasoning for each answer.
Problem 31
The cell cycle operates in the same way in all eukaryotes, from single-celled yeast to humans, and all share numerous genes whose functions are essential for the normal progression of the cycle. Discuss why you think this is the case.
Problem 32a
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Give the genotype of the cell with six chromosomes.
Problem 32b
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Align the chromosomes as they might appear at metaphase of mitosis.
Problem 32c
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Are there any alternative alignments of the chromosomes for this cell-division stage? Explain.
Problem 32d
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Separate the chromosomes and chromatids as though mitotic anaphase and telophase have taken place.
Problem 32e
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. What are the genotypes of the daughter cells?
Problem 32f
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Align the chromosomes as they might appear at metaphase I of meiosis.
Problem 32g
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Are there any alternative alignments of the chromosomes for this cell-division stage? Explain.
Problem 32h
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Separate the chromosomes as though meiotic anaphase I and telophase I have taken place.
Problem 32i
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts, to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Align the chromosomes of each daughter cell as they might appear in metaphase II of meiosis.
Problem 32j
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Are there any alternative alignments of the chromosomes for this cell-division stage? Explain.
Problem 32k
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Separate the chromosomes as though anaphase II and telophase II have taken place.
Problem 32l
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. What are the genotypes of the daughter cells?
Problem 32m
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Repeat steps (h) through (l) for the alternative alignment of chromosomes you identified in step (g).
Problem 32n
From a piece of blank paper, cut out three sets of four cigar-shaped structures (a total of 12 structures). These will represent chromatids. Be sure each member of a set of four chromatids has the same length and girth. In set one, label two chromatids 'A' and two chromatids 'a.' Cut each of these chromatids about halfway across near their midpoint and slide the two 'A' chromatids together at the cuts to form a single set of attached sister chromatids. Do the same for the 'a' chromatids. In the second set of four chromatids, label two 'B' and two 'b.' Cut and slide these together as you did for the first set, joining the 'B' chromatids together and the 'b' chromatids together. Repeat this process for the third set of chromatids, labeling them as 'D' and 'd.' You now have models for three pairs of homologous chromosomes, for a total of six chromosomes. Combining your work in steps (f) through (m), provide a written explanation of the connection between meiotic cell division and Mendel's law of independent assortment.
Problem 33a
Form a small discussion group and decide on the most likely genetic explanation for each of the following situations;
A man who has red–green color blindness and a woman who has complete color vision have a son with red–green color blindness. What are the genotypes of these three people, and how do you explain the color blindness of the son?
Problem 33b
Form a small discussion group and decide on the most likely genetic explanation for each of the following situations;
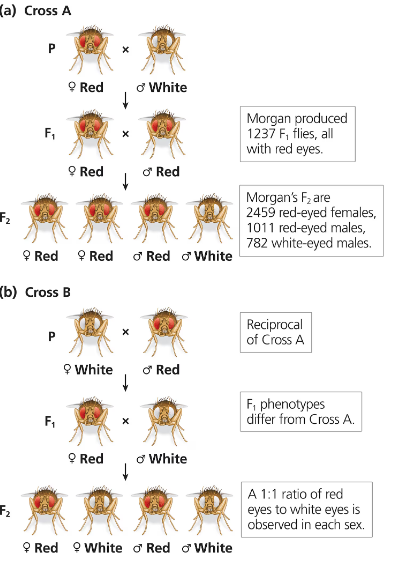
Cross A, performed by Morgan and shown in the figure below, is between a mutant male fruit fly with white eyes and a female fruit fly from a pure-breeding, red-eye stock. The figure shows that 1237 F1 progeny were produced, all of them with red eyes. In reality, this isn't entirely true. Among the 1237 F1 progeny were 3 male flies with white eyes. Give two possible explanations for the appearance of these white-eyed males.
Problem 34
Duchenne muscular dystrophy (DMD; OMIM 310200) and Becker muscular dystrophy (BMD; OMIM 300376) are both X-linked recessive conditions that result from different mutations of the same gene, known as dystrophin, on the long arm of the chromosome. BMD and DMD are quite different clinically. DMD is a very severe disorder that first appears at a young age, progresses rapidly, and is often fatal in the late teens to 20s. BMD, on the other hand, is much milder. Often symptoms don't first appear until the 40s or 50s, the progression of the disease is slow, and fatalities due to BMD are infrequent. Go to https://www.ncbi.nlm.nih/omim and survey the information describing the gene mutations causing these two conditions. Discuss the information you find with a few others in a small group, and write a single summary explaining your findings.
Problem 35
Red–green color blindness is a relatively common condition found in about 8% of males in the general population. From this, population, biologists estimate that 8% is the frequency of X chromosomes carrying a mutation of the gene encoding red and green color vision. Based on this frequency, determine the approximate frequency with which you would expect females to have red–green color blindness. Explain your reasoning.