Back
Back Sanders 3rd Edition
Sanders 3rd Edition Ch. 12 - Regulation of Gene Expression in Bacteria and Bacteriophage
Ch. 12 - Regulation of Gene Expression in Bacteria and BacteriophageProblem 26a
Suppose that base substitution mutations sufficient to eliminate the function of the operator regions listed below were to occur. For each case, describe how transcription or life cycle would be affected.
lacO mutation in E. coli
Problem 26b
Suppose that base substitution mutations sufficient to eliminate the function of the operator regions listed below were to occur. For each case, describe how transcription or life cycle would be affected.
OR1 mutation in λ phage
Problem 26c
Suppose that base substitution mutations sufficient to eliminate the function of the operator regions listed below were to occur. For each case, describe how transcription or life cycle would be affected.
OR3 mutation in λ phage
Problem 27
Two different mutations affect PRE. Mutant 1 decreases transcription from the promoter to 10% of normal. Mutant 2 increases transcription from the promoter to 10 times greater than the wild type. How will each mutation affect the determination of the lytic or lysogenic life cycle in mutant λ phage strains? Explain your answers.
Problem 28a
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
cI
Problem 28b
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
cII
Problem 28c
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
cro
Problem 28d
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
int
Problem 28e
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
cII and cro
Problem 28f
How would mutations that inactivate each of the following genes affect the determination of the lytic or lysogenic life cycle in mutated λ phage strains? Explain your answers.
N
Problem 29a
The bacterial insertion sequence IS10 uses antisense RNA to regulate translation of the mRNA that produces the enzyme transposase, which is required for insertion sequence transposition. Transcription of the antisense RNA gene is controlled by POUT, which is more than 10 times more efficient at transcription than the PIN promoter, which controls transposase gene transcription.
If a mutation reduced the transcriptional efficiency of POUT so as to be equal to that of PIN, what is the likely effect on the transposition of IS10?
Problem 29b
The bacterial insertion sequence IS10 uses antisense RNA to regulate translation of the mRNA that produces the enzyme transposase, which is required for insertion sequence transposition. Transcription of the antisense RNA gene is controlled by POUT, which is more than 10 times more efficient at transcription than the PIN promoter, which controls transposase gene transcription.
If a mutation of PIN eliminates its ability to function in transcription, what is the likely effect on the transposition of IS10?
Problem 30a
For an E. coli strain with the lac operon genotype I⁺ P⁺ O⁺ Z⁺ Y⁺, identify the level of transcription of the operon genes in each growth medium listed. Specify transcription as 'none,' 'basal,' or 'activated' for each medium, and provide an explanation to justify your answer.
Growth medium contains lactose and glucose.
Problem 30b
For an E. coli strain with the lac operon genotype I⁺ P⁺ O⁺ Z⁺ Y⁺, identify the level of transcription of the operon genes in each growth medium listed. Specify transcription as 'none,' 'basal,' or 'activated' for each medium, and provide an explanation to justify your answer.
Growth medium contains glucose but no lactose.
Problem 30c
For an E. coli strain with the lac operon genotype I⁺ P⁺ O⁺ Z⁺ Y⁺, identify the level of transcription of the operon genes in each growth medium listed. Specify transcription as 'none,' 'basal,' or 'activated' for each medium, and provide an explanation to justify your answer.
Growth medium contains lactose but no glucose.
Problem 31
How could antisense RNA be used as an antibiotic? What types of genes would you target using this scheme?
Problem 32a
The function of tRNA synthetases is to attach amino acids to tRNAs. Suppose the tRNA synthetase responsible for attaching tryptophan to tRNA is mutated in a bacterial strain, with the result that the tRNA synthetase functions at about 15% of the efficiency of the wild-type tRNA synthetase.
How would this mutation affect attenuation of the tryptophan operon? Explain your answer.
Problem 32b
The function of tRNA synthetases is to attach amino acids to tRNAs. Suppose the tRNA synthetase responsible for attaching tryptophan to tRNA is mutated in a bacterial strain, with the result that the tRNA synthetase functions at about 15% of the efficiency of the wild-type tRNA synthetase. Would formation of the 3–4 stem-loop structure in mRNA be more frequent or less frequent in the mutant strain than in the wild-type strain? Why?
Problem 33
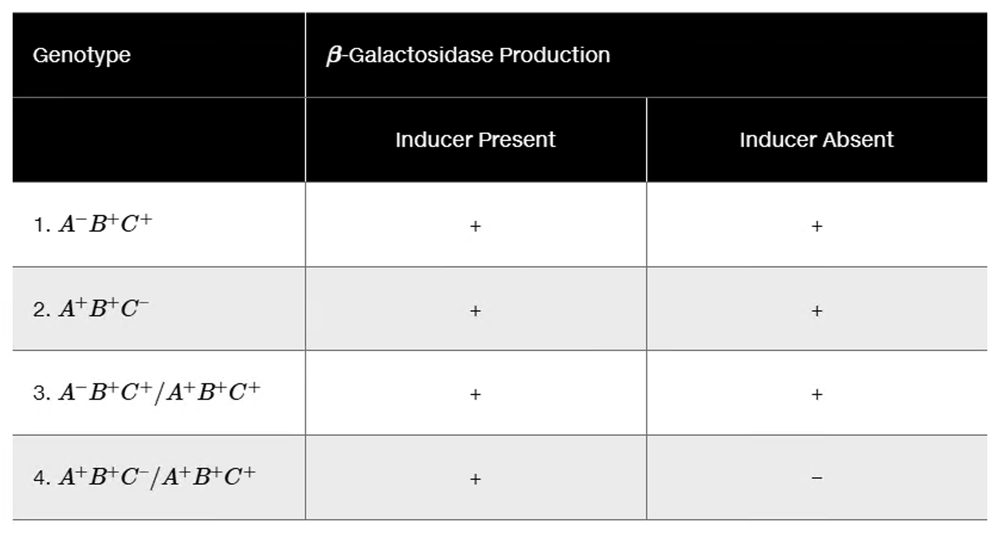
The following hypothetical genotypes have genes A, B, and C corresponding to lacI, lacO, and lacZ, but not necessarily in that order. Data in the table indicate whether β-galactosidase is produced in the presence and absence of the inducer for each genotype. Use these data to identify the correspondence between A, B, and C and the lacI, lacO, and lacZ genes. Carefully explain your reasoning for identifying each gene.
Problem 34a
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁺ bacteria with the genotype I⁺ P⁺ OC Z⁺ Y⁺
Problem 34b
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁺ O⁺ Z⁻ Y⁺
Problem 34c
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁻ OC Z⁺ Y⁺
Problem 34d
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁺ bacteria with the genotype I⁻ P⁺ OC Z⁺ Y⁺
Problem 34e
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁺ O⁺ Z⁻ Y⁺ that has a polar mutation affecting the lacZ gene
Problem 34f
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁺ OC Z⁻ Y⁻
Problem 34g
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁺ O⁺ Z⁺ Y⁺ and a mutation that prevents CAP–cAMP binding to the CAP site
Problem 35
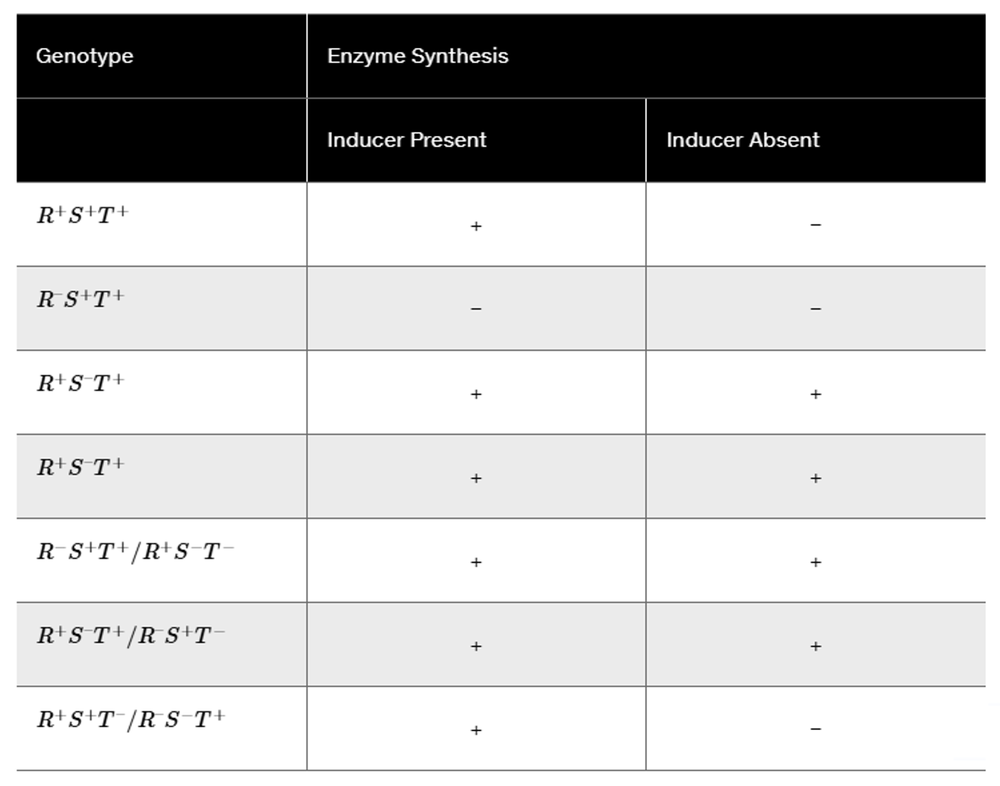
A bacterial inducible operon, similar to the lac operon, contains three genes—R, T, and S—that are involved in coordinated regulation of transcription. One of these genes is an operator region, one is a regulatory protein, and the third produces a structural enzyme. In the table below, '+' indicates that the structural enzyme is synthesized and '−' indicates that it is not produced. Use the information provided to determine which gene is the operator, which produces the regulatory protein, and which produces the enzyme.
Problem 36
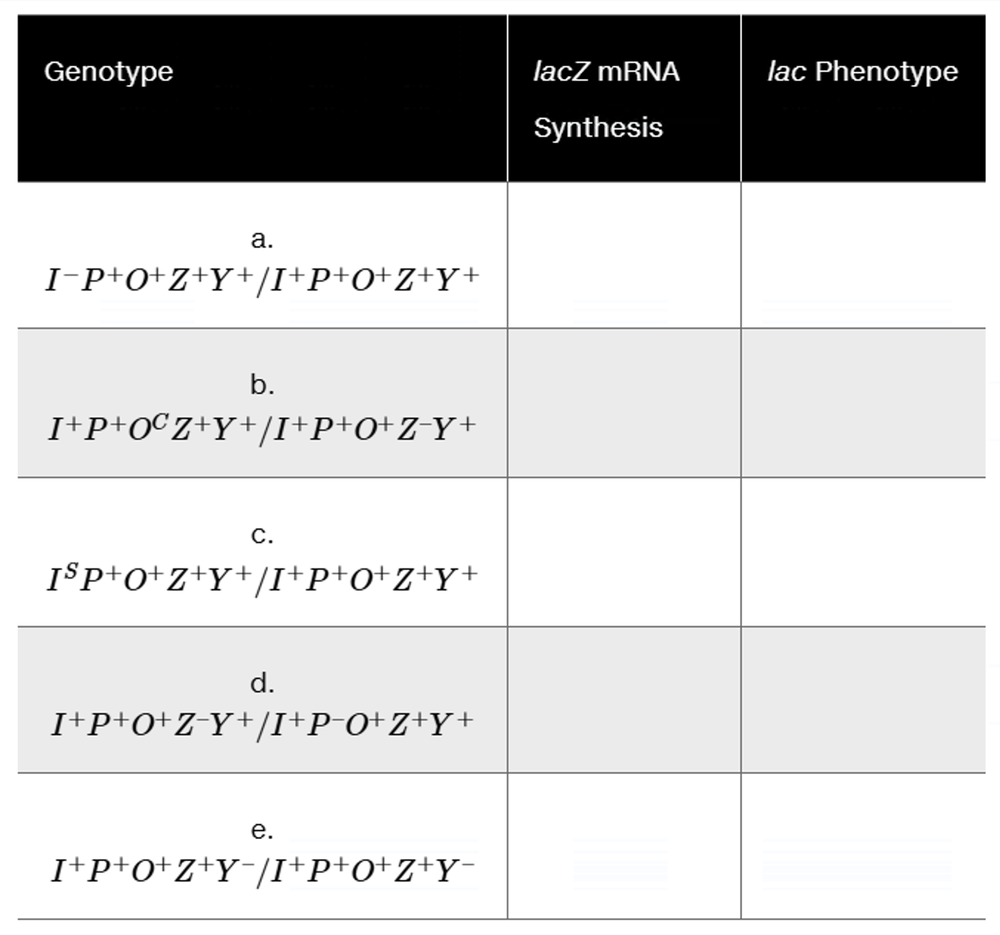
For the following lac operon partial diploids, determine whether the synthesis of lacZ mRNA is 'constitutive,' 'inducible,' or 'uninducible,' and indicate whether the partial diploid is or (able or not able to utilize lactose).
Problem 37a
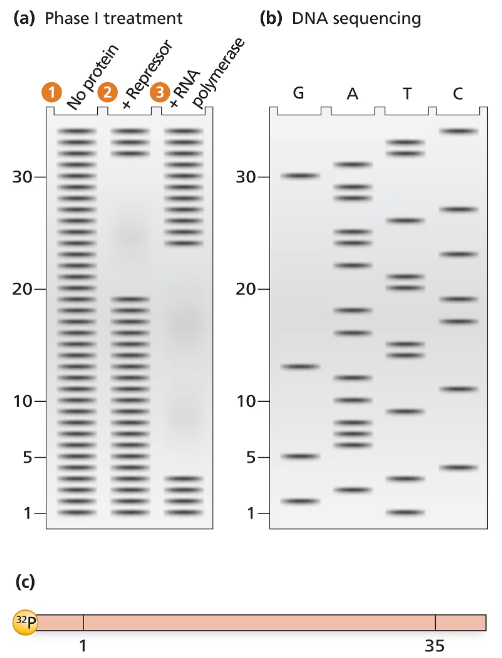
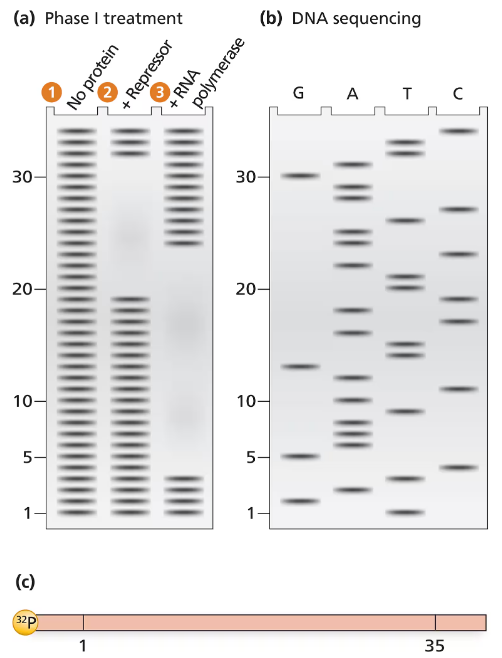
The electrophoresis gel shown in part (a) is from a DNase I footprint analysis of an operon transcription control region. DNA sequence analysis of a 35-bp region is shown in part (b). The control region, labeled with ³²P at one end, is shown in a map in part (c). Separate samples of control-region DNA are exposed to DNase I, and the resulting DNase I–digested DNA is run in separate lanes of the electrophoresis gel. Unprotected DNA is in lane 1, DNA protected by repressor protein is in lane 2, and RNA polymerase–protected DNA is in lane 3. The numbers along the electrophoresis gel correspond to the 35-bp sequence labeled on the map in part (c). Use the information provided to solve the following problems.
Determine the DNA sequence of the 35-bp region examined.
Problem 37b
The electrophoresis gel shown in part (a) is from a DNase I footprint analysis of an operon transcription control region. DNA sequence analysis of a 35-bp region is shown in part (b). The control region, labeled with ³²P at one end, is shown in a map in part (c). Separate samples of control-region DNA are exposed to DNase I, and the resulting DNase I–digested DNA is run in separate lanes of the electrophoresis gel. Unprotected DNA is in lane 1, DNA protected by repressor protein is in lane 2, and RNA polymerase–protected DNA is in lane 3. The numbers along the electrophoresis gel correspond to the 35-bp sequence labeled on the map in part (c). Use the information provided to solve the following problems.
Locate the regions of the sequence protected by repressor protein and by RNA polymerase.