Back
BackProblem 32c
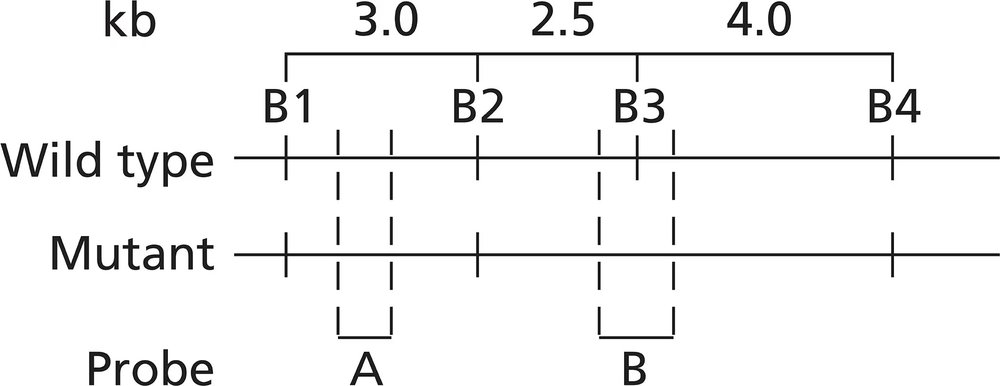
Alkaptonuria is a human autosomal recessive disorder caused by mutation of the HAO gene that encodes the enzyme homogentisic acid oxidase. A map of the HAO gene region reveals four BamHI restriction sites (B1 to B4) in the wild-type allele and three BamHI restriction sites in the mutant allele. BamHI utilizes the restriction sequence 5′-GGATCC-3′. The BamHI restriction sequence identified as B3 is altered to 5′-GGAACC-3′ in the mutant allele. The mutation results in a Ser-to-Thr missense mutation. Restriction maps of the two alleles are shown below, and the binding sites of two molecular probes (probe A and probe B) are identified.
DNA samples taken from a mother (M), father (F), and two children (C1 and C2) are analyzed by Southern blotting of BamHI-digested DNA. The gel electrophoresis results are illustrated.
Explain how the DNA sequence change results in a Ser-to-Thr missense mutation.
Problem 33a
In an experiment employing the methods of the Ames test, two strains of Salmonella are used. Strain A contains a base-substitution mutation, and Strain B contains a frameshift mutation. Four plates are prepared to test the mutagenicity of the compound ethyl methanesulfonate (EMS). Plate 1 is a control plate with Strain A and S9 extract but no EMS. Plate 2 is also a control plate and contains Strain B and S9 extract but no EMS. Plate 3 contains Strain A along with S9 extract and EMS, and Plate 4 contains Strain B, S9 extract, and EMS.
Characterize the expected distribution of colony growth on the four plates. Defend your growth prediction for each plate.
Problem 33b
In an experiment employing the methods of the Ames test, two strains of Salmonella are used. Strain A contains a base-substitution mutation, and Strain B contains a frameshift mutation. Four plates are prepared to test the mutagenicity of the compound ethyl methanesulfonate (EMS). Plate 1 is a control plate with Strain A and S9 extract but no EMS. Plate 2 is also a control plate and contains Strain B and S9 extract but no EMS. Plate 3 contains Strain A along with S9 extract and EMS, and Plate 4 contains Strain B, S9 extract, and EMS.
What event is being detected by growth of a colony on any of the four plates?
Problem 33c
In an experiment employing the methods of the Ames test, two strains of Salmonella are used. Strain A contains a base-substitution mutation, and Strain B contains a frameshift mutation. Four plates are prepared to test the mutagenicity of the compound ethyl methanesulfonate (EMS). Plate 1 is a control plate with Strain A and S9 extract but no EMS. Plate 2 is also a control plate and contains Strain B and S9 extract but no EMS. Plate 3 contains Strain A along with S9 extract and EMS, and Plate 4 contains Strain B, S9 extract, and EMS.
Why is the S9 extract added to each of the plates?
Problem 33d
In an experiment employing the methods of the Ames test, two strains of Salmonella are used. Strain A contains a base-substitution mutation, and Strain B contains a frameshift mutation. Four plates are prepared to test the mutagenicity of the compound ethyl methanesulfonate (EMS). Plate 1 is a control plate with Strain A and S9 extract but no EMS. Plate 2 is also a control plate and contains Strain B and S9 extract but no EMS. Plate 3 contains Strain A along with S9 extract and EMS, and Plate 4 contains Strain B, S9 extract, and EMS.
Suppose the compound being tested was proflavin instead of EMS. Would this change the Ames test results? Explain why or why not.
Problem 34a
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
A change in DNA sequence caused by a mistake made by DNA polymerase during replication
Problem 34b
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
Heavily damaged bacterial DNA
Problem 34c
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
A thymine dimer induced as a result of UV exposure
Problem 34d
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
A double-strand break that occurs just after replication in an actively dividing cell
Problem 34e
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
A double-stranded break that occurs during G1 and prevents completion of DNA replication
Problem 34f
Using your knowledge of DNA repair pathways, choose the pathway that would be used to repair the following types of DNA damage. Explain your reasoning.
A cytosine that has been deaminated to uracil
- Ataxia telangiectasia (OMIM 208900) is a human inherited disorder characterized by poor coordination (ataxia), red marks on the face (telangiectasia), increased sensitivity to X-rays and other radiation, and an increased susceptibility to cancer. Recent studies have shown that this disorder occurs as a result of mutation of the ATM gene. Propose a mechanism for how a mutation in the ATM gene leads to the characteristics associated with the disorder. Be sure to relate the symptoms of this disorder to functions of the ATM protein. Further, explain why DNA repair mechanisms cannot correct this problem.
Problem 35
Problem 36a
A geneticist searching for mutations uses the restriction endonucleases SmaI and PvuII to search for mutations that eliminate restriction sites. SmaI will not cleave DNA with CpG methylation. It cleaves DNA at the restriction digestion sequence ↓ 5′−CCC GGG−3′ 3′−GGG CCC−3′ ↑ PvuII is not sensitive to CpG methylation. It cleaves DNA at the restriction sequence ↓ 5′−CAG CTG−3′ 3′−GTC GAC−5′ ↑ What common feature do SmaI and PvuII share that would be useful to a researcher searching for mutations that disrupt restriction digestion?
Problem 36b
A geneticist searching for mutations uses the restriction endonucleases SmaI and PvuII to search for mutations that eliminate restriction sites. SmaI will not cleave DNA with CpG methylation. It cleaves DNA at the restriction digestion sequence ↓ 5′−CCC GGG−3′ 3′−GGG CCC−3′ ↑ PvuII is not sensitive to CpG methylation. It cleaves DNA at the restriction sequence ↓ 5′−CAG CTG−3′ 3′−GTC GAC−5′ ↑ What process is the researcher intending to detect with the use of these restriction enzymes?
Problem 36c
A geneticist searching for mutations uses the restriction endonucleases SmaI and PvuII to search for mutations that eliminate restriction sites. SmaI will not cleave DNA with CpG methylation. It cleaves DNA at the restriction digestion sequence ↓ 5′−CCC GGG−3′ 3′−GGG CCC−3′ ↑ PvuII is not sensitive to CpG methylation. It cleaves DNA at the restriction sequence ↓ 5′−CAG CTG−3′ 3′−GTC GAC−5′ ↑ Explain why CpG dinucleotides are hotspots of mutation.
Problem 37
In a mouse-breeding experiment a new mutation called Dumbo is identified. A mouse with the Dumbo mutation has very large ears. It is produced by two parental mice with normal ear size. Based on this information, can you tell whether the Dumbo mutation is a regulatory mutation or a mutation of a protein-coding gene? Why or why not?
Problem 38
Considering the Dumbo mutation in Problem 37, what kinds of additional evidence would help you determine whether Dumbo is a mutation of a regulatory sequence or of a protein-coding gene?
Problem 39
Thinking back to the discussion of gain-of-function and loss-of-function mutations, explain why gain-of-function mutations are often dominant and why loss-of-function mutations are often recessive. Give an example of a type of gain-of-function mutation that is dominant and of a loss-of-function mutation that is recessive.
Problem 40a
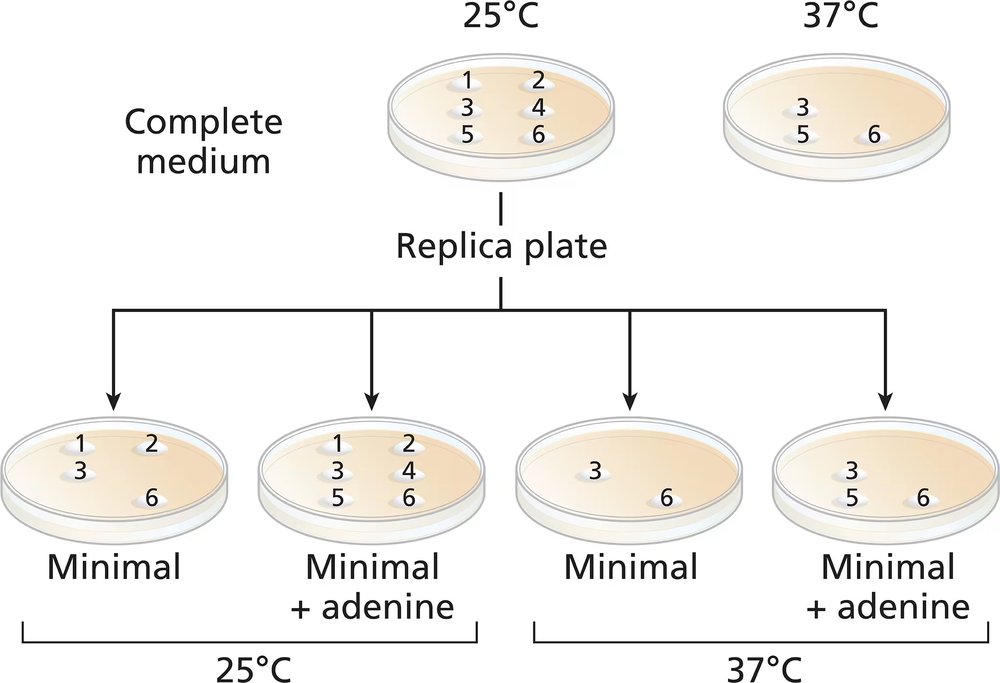
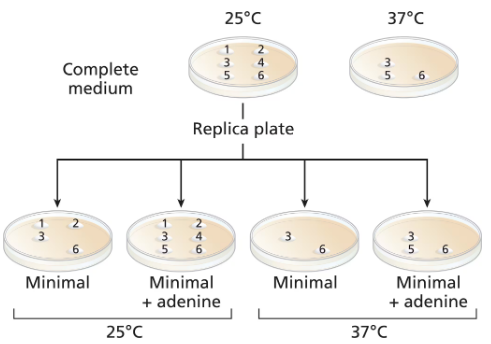
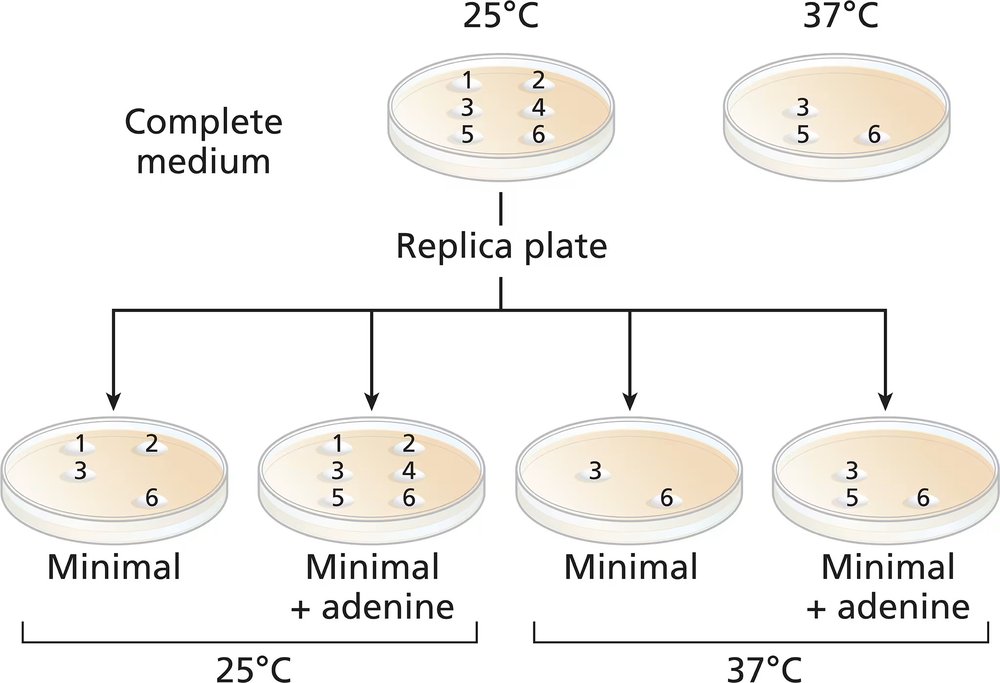
Common baker's yeast (Saccharomyces cerevisiae) is normally grown at 37°C, but it will grow actively at temperatures down to approximately 25°C. A haploid culture of wild-type yeast is mutagenized with EMS. Cells from the mutagenized culture are spread on a complete-medium plate and grown at 25°C. Six colonies (1 to 6) are selected from the original complete-medium plate and transferred to two fresh complete-medium plates. The new complete plates (shown) are grown at 25°C and 37°C. Four replica plates are made onto minimal medium or minimal plus adenine from the 25°C complete-medium plate. The new plates are grown at either 25°C or 37°C and the growth results are shown.
Which colonies are prototrophic and which are auxotrophic? What growth information is used to make these determinations?
Problem 40b
Common baker's yeast (Saccharomyces cerevisiae) is normally grown at 37°C, but it will grow actively at temperatures down to approximately 25°C. A haploid culture of wild-type yeast is mutagenized with EMS. Cells from the mutagenized culture are spread on a complete-medium plate and grown at 25°C. Six colonies (1 to 6) are selected from the original complete-medium plate and transferred to two fresh complete-medium plates. The new complete plates (shown) are grown at 25°C and 37°C. Four replica plates are made onto minimal medium or minimal plus adenine from the 25°C complete-medium plate. The new plates are grown at either 25°C or 37°C and the growth results are shown.
Classify the nature of the mutations in colonies 1, 2, and 5.
Problem 40c
Common baker's yeast (Saccharomyces cerevisiae) is normally grown at 37°C, but it will grow actively at temperatures down to approximately 25°C. A haploid culture of wild-type yeast is mutagenized with EMS. Cells from the mutagenized culture are spread on a complete-medium plate and grown at 25°C. Six colonies (1 to 6) are selected from the original complete-medium plate and transferred to two fresh complete-medium plates. The new complete plates (shown) are grown at 25°C and 37°C. Four replica plates are made onto minimal medium or minimal plus adenine from the 25°C complete-medium plate. The new plates are grown at either 25°C or 37°C and the growth results are shown.
What can you say about colony 4?
Problem 41a
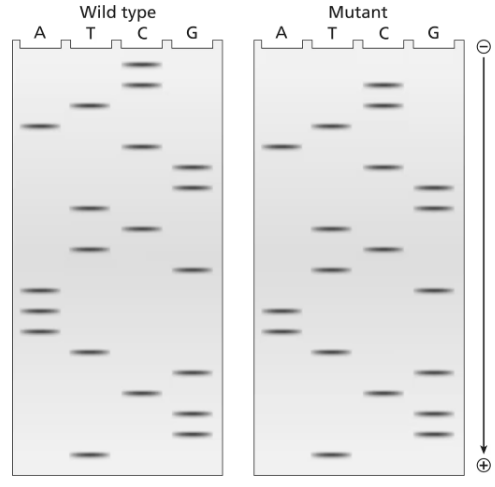
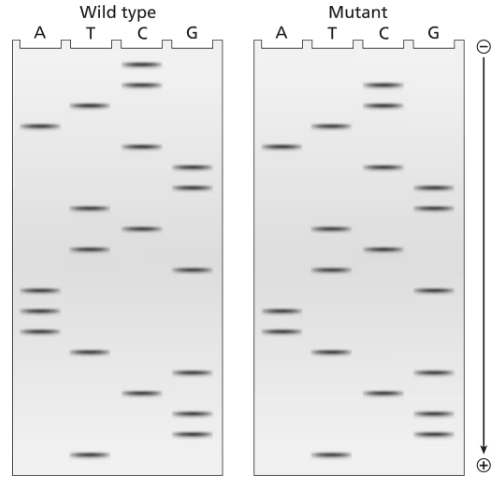
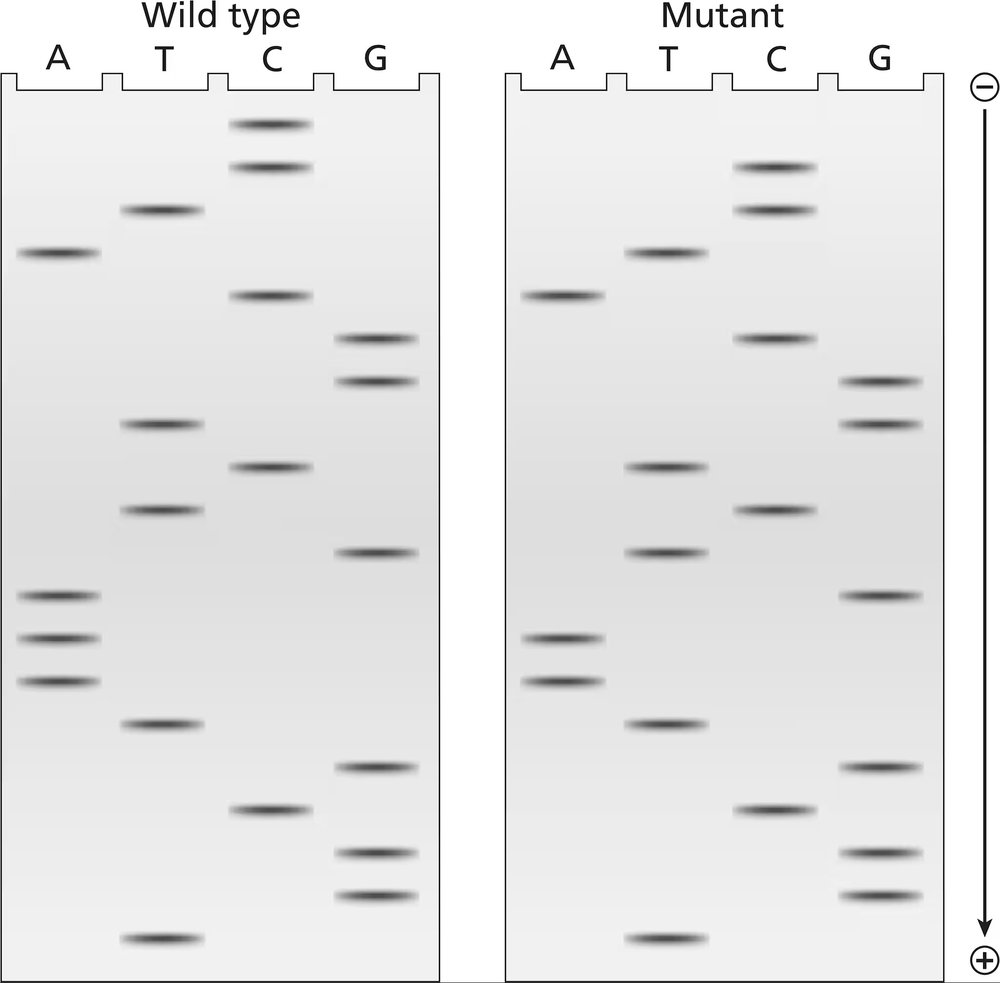
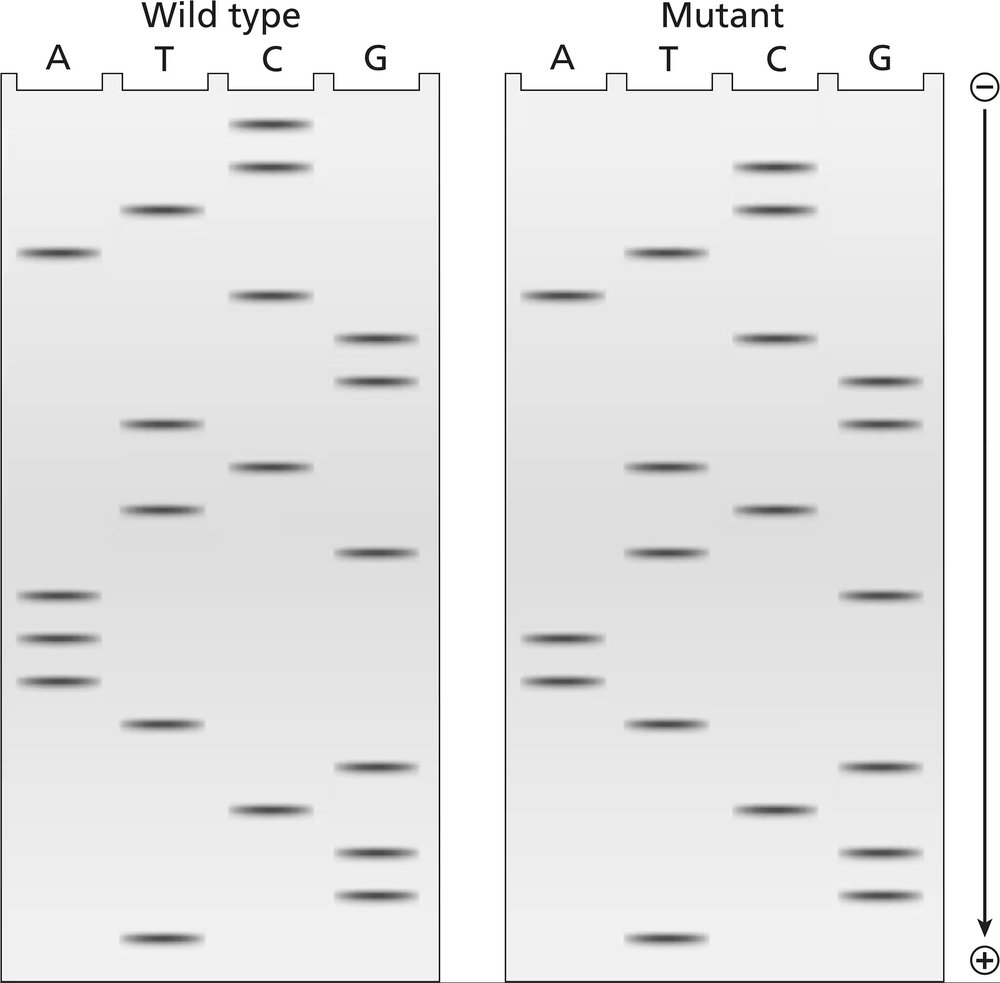
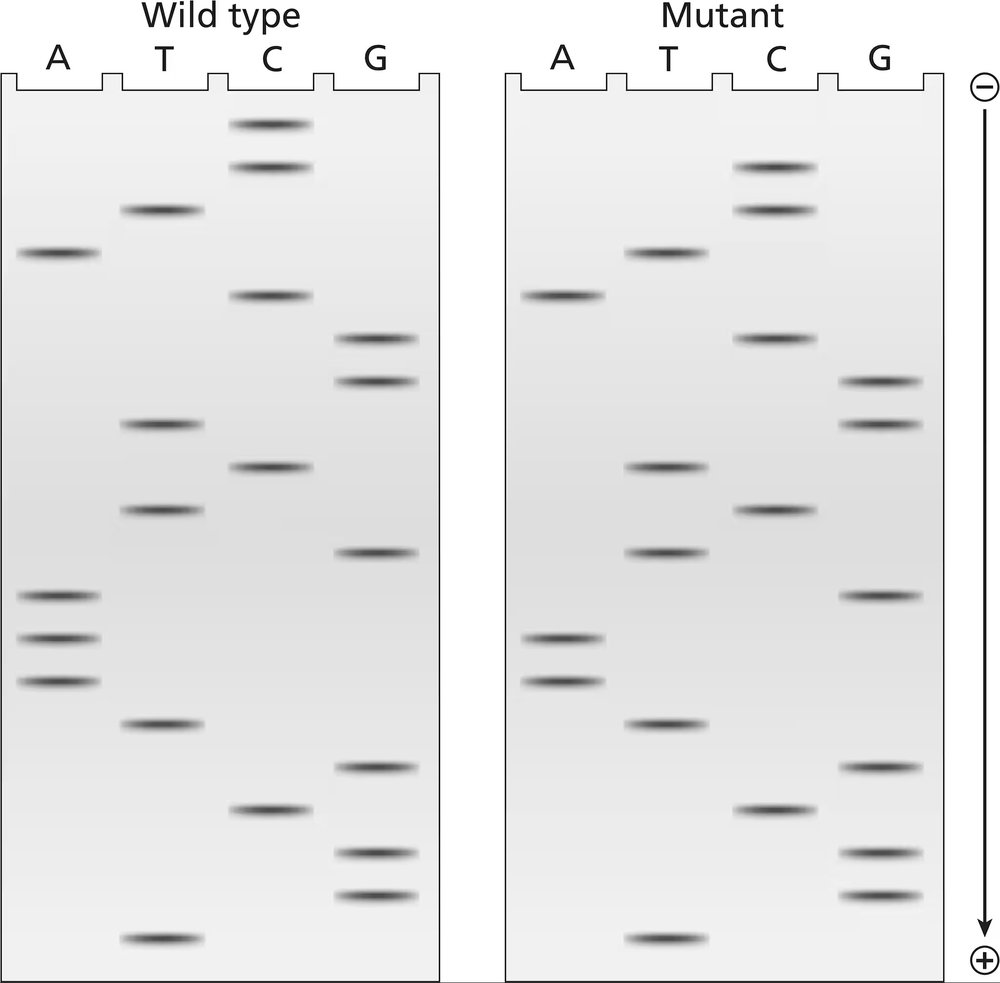
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
Write the DNA sequence of both alleles, including strand polarity.
Problem 41b
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
Identify the template and nontemplate strands of DNA.
Problem 41c
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
Write out the mRNA sequences encoded by each template strand, and underline the start codons.
Problem 41d
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
Determine the amino acid sequences translated from these mRNAs.
Problem 41e
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
What is the cause of the mutation?