Back
BackProblem 14e
Several types of mutation are identified and described in the chapter. These include (1) promoter mutation, (2) splice site mutation, (3) missense mutation, (4) frameshift mutation, and 5) nonsense mutation. Match the following mutation descriptions with the type(s) of mutations listed above. More than one mutation type might match a description.
A null mutation that does not produce any functional protein product.
Problem 15
A 1-mL sample of the bacterium E. coli is exposed to ultraviolet light. The sample is used to inoculate a 500-mL flask of complete medium that allows growth of all bacterial cells. The 500-mL culture is grown on the benchtop, and two equal-sized samples are removed and plated on identical complete-medium growth plates. Plate 1 is immediately wrapped in a dark cloth, but plate 2 is not covered. Both plates are left at room temperature for 36 hours and then examined. Plate 2 is seen to contain many more growing colonies than plate 1.
Thinking about DNA repair processes, how do you explain this observation?
Problem 16
A strain of E. coli is identified as having a null mutation of the RecA gene. What biological property do you expect to be absent in the mutant strain? What is the molecular basis for the missing property?
Problem 17
Describe the difference between DNA transposons and retrotransposons.
Problem 18
How are flanking direct repeat sequences created by transposition?
Problem 19a
Using the adenine–thymine base pair in this DNA sequence
...GCTC...
...CGAG...
Give the sequence after a transition mutation.
Problem 19b
Using the adenine–thymine base pair in this DNA sequence
...GCTC...
...CGAG...
Give the sequence after a transversion mutation.
Problem 20a
The partial amino acid sequence of a wild-type protein is
… Arg-Met-Tyr-Thr-Leu-Cys-Ser …
The same portion of the protein from a mutant has the sequence
… Arg-Met-Leu-Tyr-Ala-Leu-Phe …
Identify the type of mutation.
Problem 20b
The partial amino acid sequence of a wild-type protein is
… Arg-Met-Tyr-Thr-Leu-Cys-Ser …
The same portion of the protein from a mutant has the sequence
… Arg-Met-Leu-Tyr-Ala-Leu-Phe …
Give the sequence of the wild-type DNA template strand. Use A/G if the nucleotide could be either purine, T/C if it could be either pyrimidine, N if any nucleotide could occur at a site, or the alternative nucleotides if a purine and a pyrimidine are possible.
Problem 21
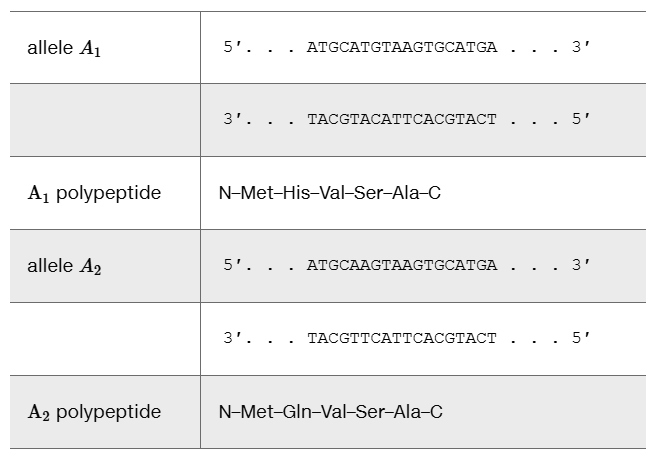
The two DNA and polypeptide sequences shown are for alleles at a hypothetical locus that produce different polypeptides, both five amino acids long. In each case, the lower DNA strand is the template strand:
Based on DNA and polypeptide sequences alone, is there any way to determine which allele is dominant and which is recessive? Why or why not?
Problem 22a
Many human genes are known to have homologs in the mouse genome. One approach to investigating human hereditary disease is to produce mutations of the mouse homologs of human genes by methods that can precisely target specific nucleotides for mutation.
Numerous studies of mutations of the mouse homologs of human genes have yielded valuable information about how gene mutations influence the human disease process. In general terms, describe how and why creating mutations of the mouse homologs can give information about human hereditary disease processes.
Problem 22b
Many human genes are known to have homologs in the mouse genome. One approach to investigating human hereditary disease is to produce mutations of the mouse homologs of human genes by methods that can precisely target specific nucleotides for mutation.
Despite the homologies that exist between human and mouse genes, some attempts to study human hereditary disease processes by inducing mutations in mouse genes indicate there is little to be learned about human disease in this way. In general terms, describe how and why the study of mouse gene mutations might fail to produce useful information about human disease processes.
- The fluctuation test performed by Luria and Delbrück is consistent with the random mutation hypothesis. Briefly describe their experiment and identify how the results match the prediction of the random mutation hypothesis. What would have to be different about the experimental results for them to agree with the prediction of the adaptive mutation hypothesis?
Problem 23
- In this chapter, three features of genes or of DNA sequence that contribute to the occurrence of mutational hotspots were described. Identify those three features and briefly describe why they are associated with mutational hotspots.
Problem 24
- Briefly compare the production of DNA double-strand breaks in bacteria versus the double-strand breaks that precede homologous recombination.
Problem 25
- During mismatch repair, why is it necessary to distinguish between the template strand and the newly made daughter strand? Describe how this is accomplished.
Problem 26
Problem 27
Following the spill of a mixture of chemicals into a small pond, bacteria from the pond are tested and show an unusually high rate of mutation. A number of mutant cultures are grown from mutant colonies and treated with known mutagens to study the rate of reversion. Most of the mutant cultures show a significantly higher reversion rate when exposed to base analogs such as proflavin and 2-aminopurine. What does this suggest about the nature of the chemicals in the spill?
Problem 28a
In an Ames test using his⁻ Salmonella bacteria a researcher determines that adding a test compound plus the S9 extract produces a large number of his⁺ revertants but mixing the his⁻ strain plus the test compound without adding S9 does not produce an elevated number of his⁺ revertants.
What is the reason for the different experimental results described?
Problem 28b
In an Ames test using his⁻ Salmonella bacteria a researcher determines that adding a test compound plus the S9 extract produces a large number of his⁺ revertants but mixing the his⁻ strain plus the test compound without adding S9 does not produce an elevated number of his⁺ revertants.
Is the test compound still considered to be a potential mutagen? Explain why or why not.
Problem 29a
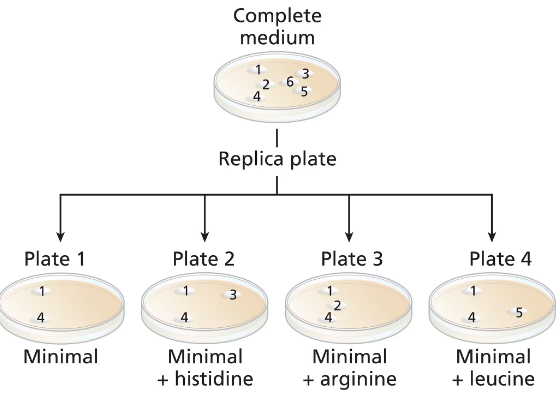
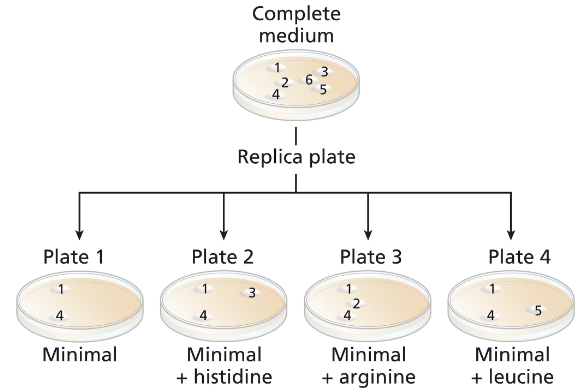
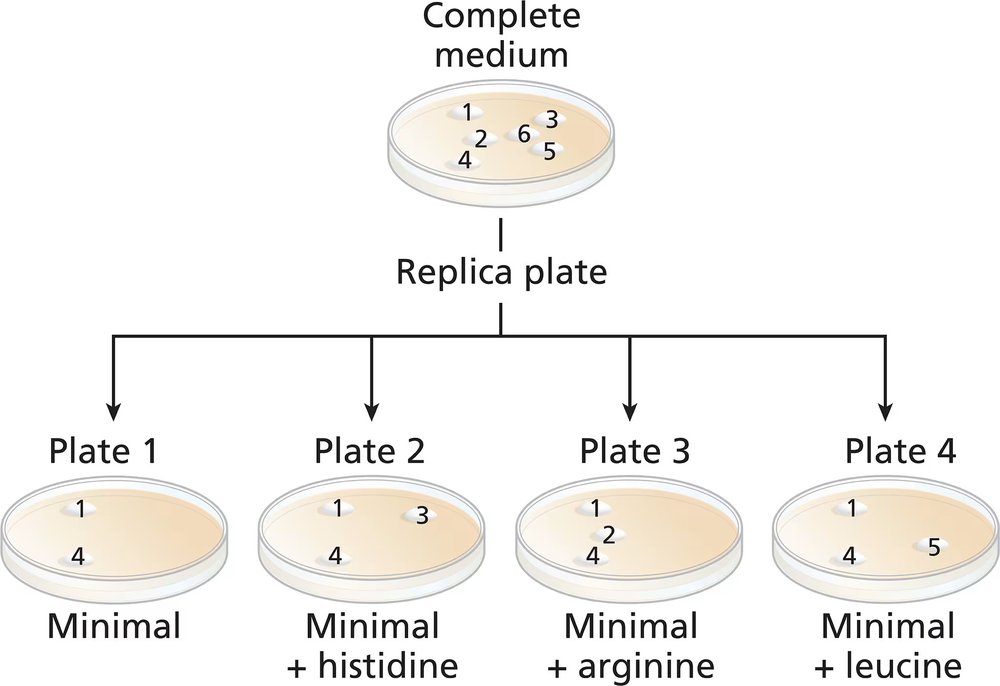
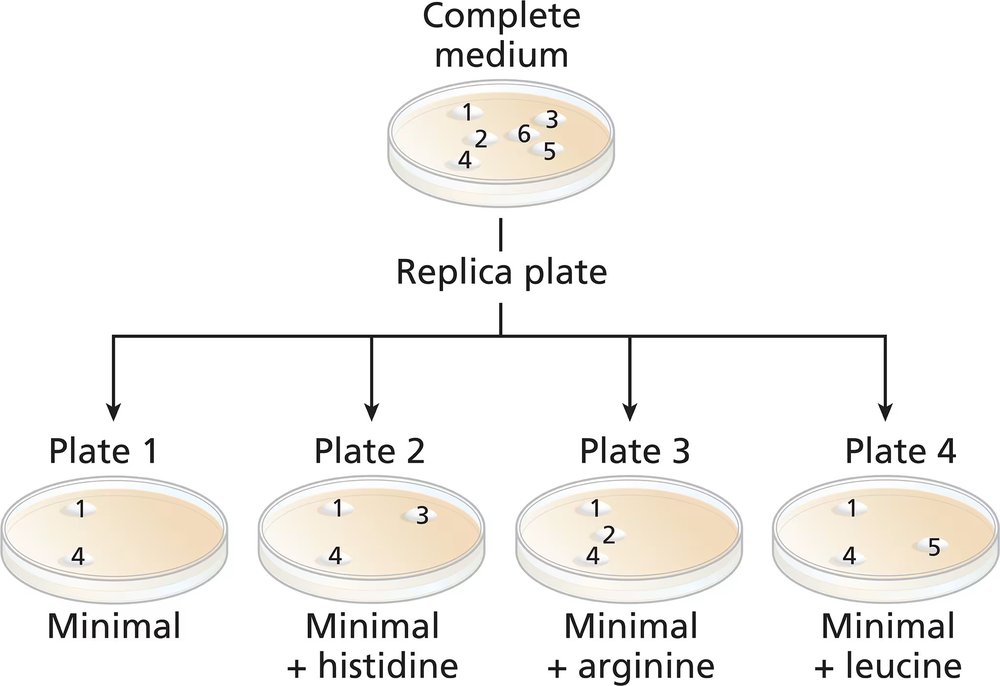
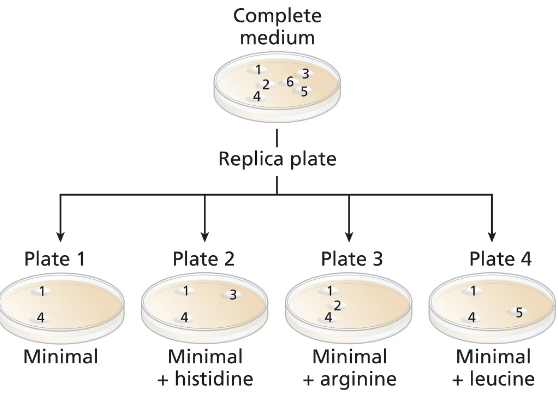
A wild-type culture of haploid yeast is exposed to ethyl methanesulfonate (EMS). Yeast cells are plated on a complete medium, and 6 colonies (colonies numbered 1 to 6) are transferred to a new complete medium plate for further study. Four replica plates are made from the complete medium plate to plates containing minimal medium or minimal medium plus one amino acid (replica plates numbered 1 to 4) with the following results:
Identify the colonies that are prototrophic (wild type). What growth information leads to your answer?
Problem 29b
A wild-type culture of haploid yeast is exposed to ethyl methanesulfonate (EMS). Yeast cells are plated on a complete medium, and 6 colonies (colonies numbered 1 to 6) are transferred to a new complete medium plate for further study. Four replica plates are made from the complete medium plate to plates containing minimal medium or minimal medium plus one amino acid (replica plates numbered 1 to 4) with the following results:
Identify the colonies that are auxotrophic (mutant). What growth information leads to your answer?
Problem 29c
A wild-type culture of haploid yeast is exposed to ethyl methanesulfonate (EMS). Yeast cells are plated on a complete medium, and 6 colonies (colonies numbered 1 to 6) are transferred to a new complete medium plate for further study. Four replica plates are made from the complete medium plate to plates containing minimal medium or minimal medium plus one amino acid (replica plates numbered 1 to 4) with the following results:
Identify any colonies that are his⁻, arg⁻, leu⁻.
Problem 29d
A wild-type culture of haploid yeast is exposed to ethyl methanesulfonate (EMS). Yeast cells are plated on a complete medium, and 6 colonies (colonies numbered 1 to 6) are transferred to a new complete medium plate for further study. Four replica plates are made from the complete medium plate to plates containing minimal medium or minimal medium plus one amino acid (replica plates numbered 1 to 4) with the following results:
For colonies 1, 3, and 5, write '+' for the wild-type synthesis and '−' for the mutant synthesis of histidine and leucine.
Problem 29e
A wild-type culture of haploid yeast is exposed to ethyl methanesulfonate (EMS). Yeast cells are plated on a complete medium, and 6 colonies (colonies numbered 1 to 6) are transferred to a new complete medium plate for further study. Four replica plates are made from the complete medium plate to plates containing minimal medium or minimal medium plus one amino acid (replica plates numbered 1 to 4) with the following results:
Are there any colonies for which genotype information cannot be determined? If so, which colony or colonies?
Problem 30a
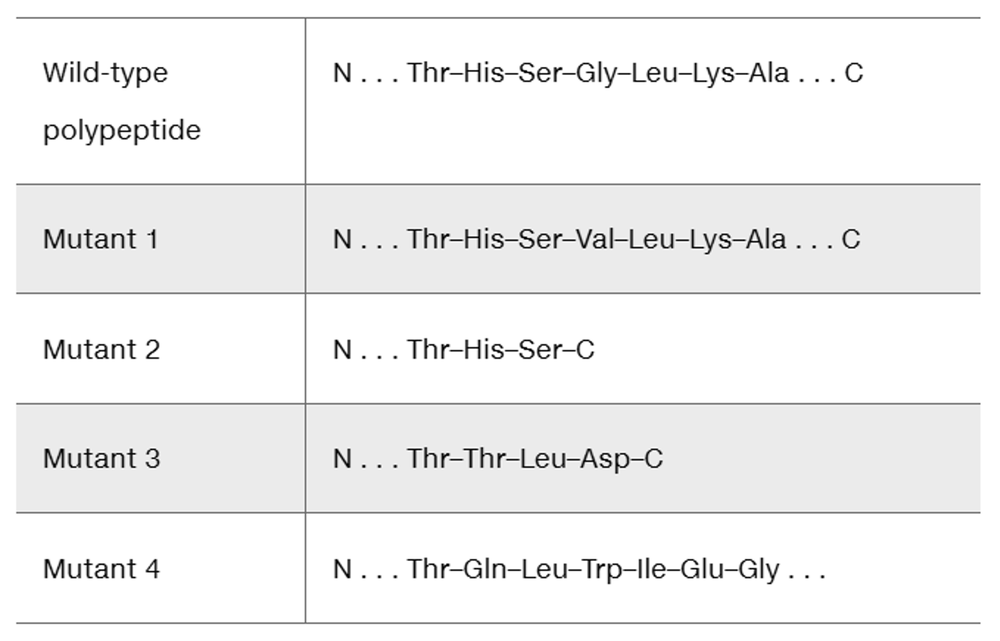
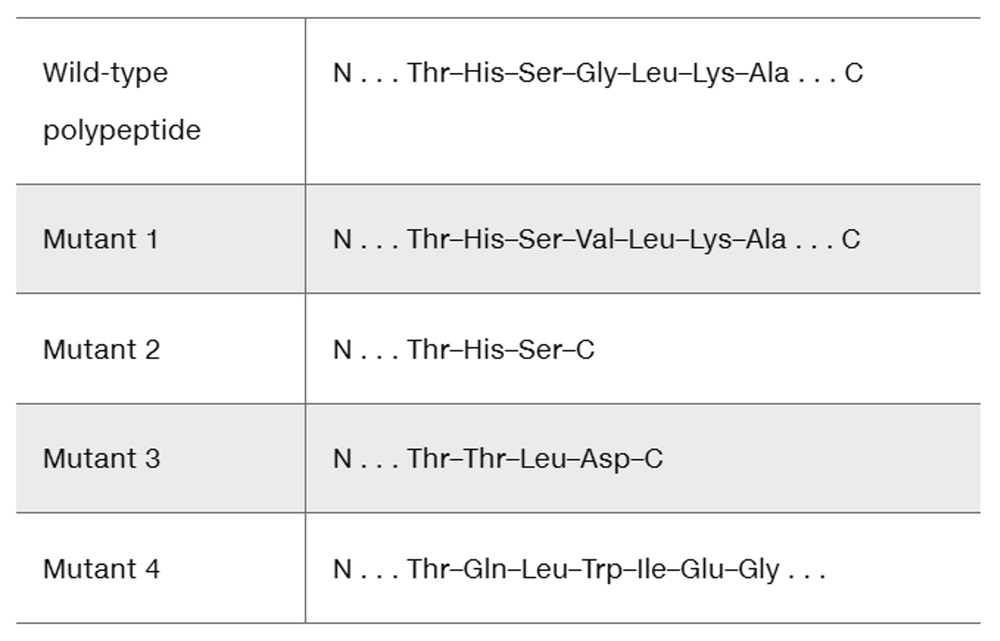
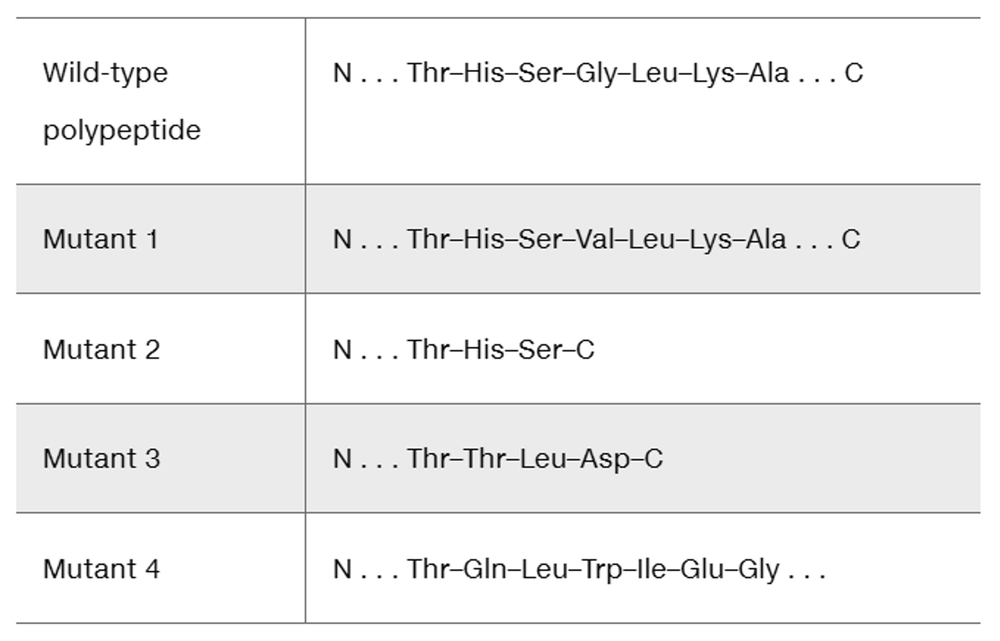
A fragment of a wild-type polypeptide is sequenced for seven amino acids. The same polypeptide region is sequenced in four mutants.
Use the available information to characterize each mutant.
Problem 30b
A fragment of a wild-type polypeptide is sequenced for seven amino acids. The same polypeptide region is sequenced in four mutants.
Determine the wild-type mRNA sequence.
Problem 30c
A fragment of a wild-type polypeptide is sequenced for seven amino acids. The same polypeptide region is sequenced in four mutants.
Identify the mutation that produces each mutant polypeptide.
Problem 31
Experiments by Charles Yanofsky in the 1950s and 1960s helped characterize the nature of tryptophan synthesis in E. coli. In one of Yanofsky's experiments, he identified glycine (Gly) as the wild-type amino acid in position 211 of tryptophan synthetase, the product of the trpA gene. He identified two independent missense mutants with defective tryptophan synthetase at these positions that resulted from base-pair substitutions. One mutant encoded arginine (Arg) and another encoded glutamic acid (Glu). At position 235, wild-type tryptophan synthetase contains serine (Ser) but a base-pair substitution mutant encodes leucine (Leu). At position 243, the wild-type polypeptide contains glutamine and a base-pair substitution mutant encodes a stop codon. Identify the most likely wild-type codons for positions 211, 235, and 243. Justify your answer in each case.
Problem 32a
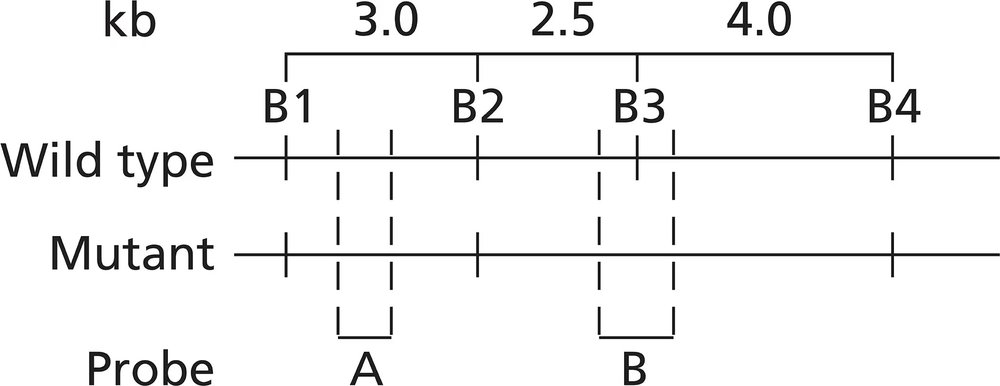
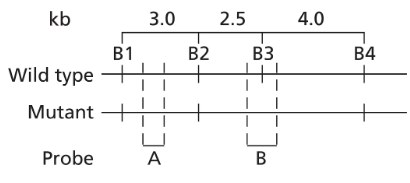
Alkaptonuria is a human autosomal recessive disorder caused by mutation of the HAO gene that encodes the enzyme homogentisic acid oxidase. A map of the HAO gene region reveals four BamHI restriction sites (B1 to B4) in the wild-type allele and three BamHI restriction sites in the mutant allele. BamHI utilizes the restriction sequence 5′-GGATCC-3′. The BamHI restriction sequence identified as B3 is altered to 5′-GGAACC-3′ in the mutant allele. The mutation results in a Ser-to-Thr missense mutation. Restriction maps of the two alleles are shown below, and the binding sites of two molecular probes (probe A and probe B) are identified.
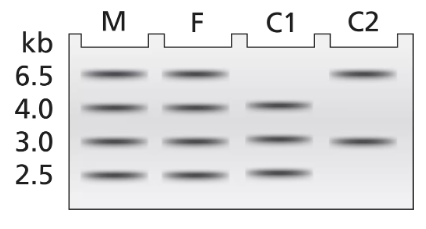
DNA samples taken from a mother (M), father (F), and two children (C1 and C2) are analyzed by Southern blotting of BamHI-digested DNA. The gel electrophoresis results are illustrated.
Using A to represent the wild-type allele and a for the mutant allele, identify the genotype of each family member. Identify any family member who is alkaptonuric.
Problem 32b
Alkaptonuria is a human autosomal recessive disorder caused by mutation of the HAO gene that encodes the enzyme homogentisic acid oxidase. A map of the HAO gene region reveals four BamHI restriction sites (B1 to B4) in the wild-type allele and three BamHI restriction sites in the mutant allele. BamHI utilizes the restriction sequence 5′-GGATCC-3′. The BamHI restriction sequence identified as B3 is altered to 5′-GGAACC-3′ in the mutant allele. The mutation results in a Ser-to-Thr missense mutation. Restriction maps of the two alleles are shown below, and the binding sites of two molecular probes (probe A and probe B) are identified.
DNA samples taken from a mother (M), father (F), and two children (C1 and C2) are analyzed by Southern blotting of BamHI-digested DNA. The gel electrophoresis results are illustrated.
In a separate figure, draw the gel electrophoresis band patterns for all the genotypes that could be found in children of this couple.