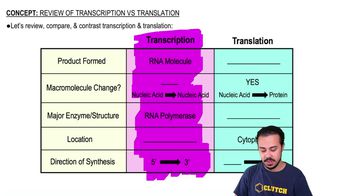
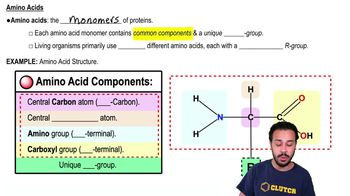
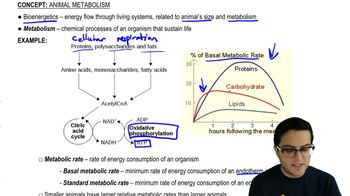
In a particular bacterial species, temperature-sensitive conditional mutations cause expression of a wild-type phenotype at one growth temperature and a mutant phenotype at another—typically higher—temperature. Imagine that when a bacterial cell carrying such a mutation is shifted from low to high growth temperatures, RNA polymerases in the process of elongation complete transcription normally, but no new transcripts can be started. The mutation in this strain most likely affects:
a. The terminator sequence
b. The start codon
c. Sigma
d. One of the polypeptides of the core RNA polymerase