Back
BackProblem 10
What is the role of enhancer sequences in transcription of eukaryotic genes? Speculate about why enhancers are not part of transcription of bacterial genes.
Problem 11
Describe the difference between introns and exons.
Problem 12
Draw a bacterial promoter and label its consensus sequences. How does this promoter differ from a eukaryotic promoter transcribed:
By RNA polymerase II?
By RNA polymerase I?
By RNA polymerase III?
Problem 13
For a eukaryotic gene whose transcription requires the activity of an enhancer sequence, explain how proteins bound at the enhancer interact with RNA pol II and transcription factors bound at the promoter.
Problem 14
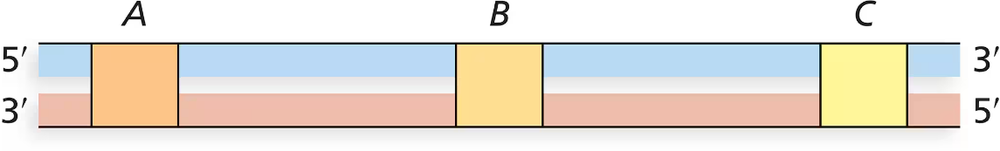
Three genes identified in the diagram as A, B, and C are transcribed from a region of DNA. The 5'-to-3' transcription of genes A and C elongates mRNA in the right-to-left direction, and transcription of gene B elongates mRNA in the left-to-right direction. For each gene, identify the coding strand by designating it as an 'upper strand' or 'lower strand' in the diagram.
Problem 15a
The eukaryotic gene Gen-100 contains four introns labeled A to D. Imagine that Gen-100 has been isolated and its DNA has been denatured and mixed with polyadenylated mRNA from the gene.
Illustrate the R-loop structure that would be seen with electron microscopy.
Problem 15b
The eukaryotic gene Gen-100 contains four introns labeled A to D. Imagine that Gen-100 has been isolated and its DNA has been denatured and mixed with polyadenylated mRNA from the gene.
Label the introns.
Problem 15c
The eukaryotic gene Gen-100 contains four introns labeled A to D. Imagine that Gen-100 has been isolated and its DNA has been denatured and mixed with polyadenylated mRNA from the gene.
Are intron regions single stranded or double stranded? Why?
Problem 16a
The segment of the bacterial TrpA gene involved in intrinsic termination of transcription is the following:
3'-TGGGTCGGGGCGGATTACTGCCCCGAAAAAAAACTTG-5'
5'-ACCCAGCCCCGCCTAATGACGGGGCTTTTTTTTGAAC-3' Draw the mRNA structure that forms during transcription of this segment of the TrpA gene.
Problem 16b
The segment of the bacterial TrpA gene involved in intrinsic termination of transcription is the following:
3'-TGGGTCGGGGCGGATTACTGCCCCGAAAAAAAACTTG-5'
5'-ACCCAGCCCCGCCTAATGACGGGGCTTTTTTTTGAAC-3'
Label the template and coding DNA strands.
Problem 16c
The segment of the bacterial TrpA gene involved in intrinsic termination of transcription is the following:
3'-TGGGTCGGGGCGGATTACTGCCCCGAAAAAAAACTTG-5'
5'-ACCCAGCCCCGCCTAATGACGGGGCTTTTTTTTGAAC-3'
Explain how a sequence of this type leads to intrinsic termination of transcription.
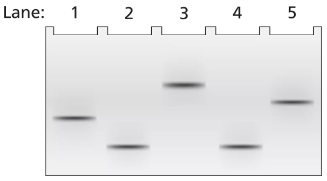
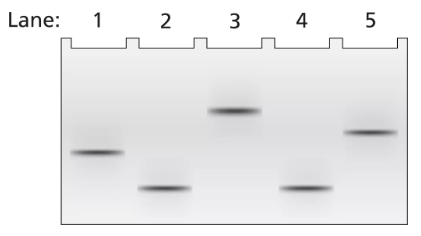
Problem 17a
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Diagram the relative positions expected for the DNA fragments in this gel electrophoresis analysis.
Problem 17b
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Explain the relative positions of bands in lanes 1 and 3.
Problem 17c
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Explain the relative positions of bands in lanes 1 and 4.
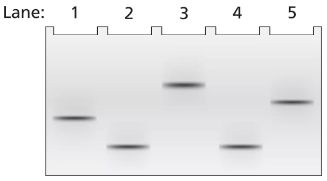
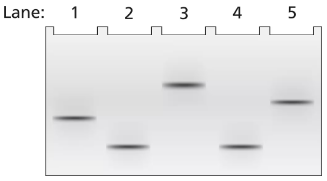
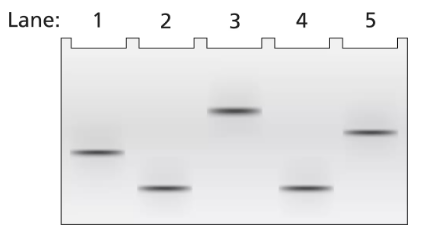
Problem 18a
A 3.5-kb segment of DNA containing the complete sequence of a mouse gene is available. The DNA segment contains the promoter sequence and extends beyond the polyadenylation site of the gene. The DNA is studied by band shift assay, and the following gel bands are observed.
Match these conditions to a specific lane of the gel.
3.5-kb fragment plus TFIIB and TFIID
Problem 18b
A 3.5-kb segment of DNA containing the complete sequence of a mouse gene is available. The DNA segment contains the promoter sequence and extends beyond the polyadenylation site of the gene. The DNA is studied by band shift assay, and the following gel bands are observed.
Match these conditions to a specific lane of the gel.
3.5-kb fragment plus TFIIB, TFIID, TFIIF, and RNA polymerase II
Problem 18c
A 3.5-kb segment of DNA containing the complete sequence of a mouse gene is available. The DNA segment contains the promoter sequence and extends beyond the polyadenylation site of the gene. The DNA is studied by band shift assay, and the following gel bands are observed.
Match these conditions to a specific lane of the gel.
3.5-kb fragment alone
Problem 18d
A 3.5-kb segment of DNA containing the complete sequence of a mouse gene is available. The DNA segment contains the promoter sequence and extends beyond the polyadenylation site of the gene. The DNA is studied by band shift assay, and the following gel bands are observed.
Match these conditions to a specific lane of the gel.
3.5-kb fragment plus RNA polymerase II
Problem 18e
A 3.5-kb segment of DNA containing the complete sequence of a mouse gene is available. The DNA segment contains the promoter sequence and extends beyond the polyadenylation site of the gene. The DNA is studied by band shift assay, and the following gel bands are observed.
Match these conditions to a specific lane of the gel.
3.5-kb fragment plus TFIIB
Problem 19a
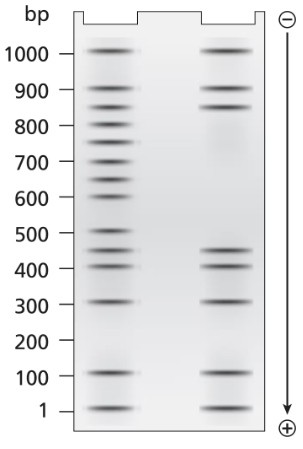
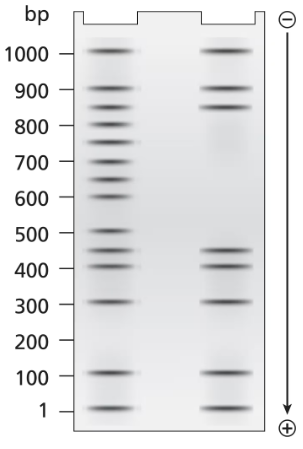
A 1.0-kb DNA fragment from the end of the mouse gene described in the previous problem is examined by DNA footprint protection analysis. Two samples are end-labeled with ³²P and one of the two is mixed with TFIIB, TFIID, and RNA polymerase II. The DNA exposed to these proteins is run in the right-hand lane of the gel shown below and the control DNA is run in the left-hand. Both DNA samples are treated with DNase I before running the samples on the electrophoresis gel.
What length of DNA is bound by the transcriptional proteins? Explain how the gel results support this interpretation.
Problem 19b
A 1.0-kb DNA fragment from the end of the mouse gene described in the previous problem is examined by DNA footprint protection analysis. Two samples are end-labeled with ³²P, and one of the two is mixed with TFIIB, TFIID, and RNA polymerase II. The DNA exposed to these proteins is run in the right-hand lane of the gel shown below and the control DNA is run in the left-hand. Both DNA samples are treated with DNase I before running the samples on the electrophoresis gel.
Draw a diagram of this DNA fragment bound by the transcriptional proteins, showing the approximate position of proteins along the fragment.
Problem 19c
A 1.0-kb DNA fragment from the end of the mouse gene described in the previous problem is examined by DNA footprint protection analysis. Two samples are end-labeled with ³²P and one of the two is mixed with TFIIB, TFIID, and RNA polymerase II. The DNA exposed to these proteins is run in the right-hand lane of the gel shown below and the control DNA is run in the left-hand. Both DNA samples are treated with DNase I before running the samples on the electrophoresis gel.
Explain the role of DNase I.
Problem 20a
Wild-type E. coli grow best at 37°C but can grow efficiently up to 42°C. An E. coli strain has a mutation of the sigma subunit that results in an RNA polymerase holoenzyme that is stable and transcribes at wild-type levels at 37°C. The mutant holoenzyme is progressively destabilized as the temperature is raised, and it completely denatures and ceases to carry out transcription at 42°C. Relative to wild-type growth, characterize the ability of the mutant strain to carry out transcription at 37°C
Problem 20b
Wild-type E. coli grows best at 37°C but can grow efficiently up to 42°C. An E. coli strain has a mutation of the sigma subunit that results in an RNA polymerase holoenzyme that is stable and transcribes at wild-type levels at 37°C. The mutant holoenzyme is progressively destabilized as the temperature is raised, and it completely denatures and ceases to carry out transcription at 42°C. Relative to wild-type growth, characterize the ability of the mutant strain to carry out transcription at 40°C
Problem 20c
Wild-type E. coli grows best at 37°C but can grow efficiently up to 42°C. An E. coli strain has a mutation of the sigma subunit that results in an RNA polymerase holoenzyme that is stable and transcribes at wild-type levels at 37°C. The mutant holoenzyme is progressively destabilized as the temperature is raised, and it completely denatures and ceases to carry out transcription at 42°C. Relative to wild-type growth, characterize the ability of the mutant strain to carry out transcription at 42°C
Problem 20d
Wild-type E. coli grows best at 37°C but can grow efficiently up to 42°C. An E. coli strain has a mutation of the sigma subunit that results in an RNA polymerase holoenzyme that is stable and transcribes at wild-type levels at 37°C. The mutant holoenzyme is progressively destabilized as the temperature is raised, and it completely denatures and ceases to carry out transcription at 42°C. Relative to wild-type growth, characterize the ability of the mutant strain to carry out transcription at What term best characterizes the type of mutation exhibited by the mutant bacterial strain? (Hint: The term was used in Chapter 4 to describe the Himalayan allele of the mammalian C gene.)
Problem 21a
A mutant strain of Salmonella bacteria carries a mutation of the rho protein that has full activity at 37°C but is completely inactivated when the mutant strain is grown at 40°C. Speculate about the kind of differences you would expect to see if you compared a broad spectrum of mRNAs from the mutant strain grown at 37°C and the same spectrum of mRNAs from the strain when grown at 40°C.
Problem 21b
A mutant strain of Salmonella bacteria carries a mutation of the rho protein that has full activity at 37°C but is completely inactivated when the mutant strain is grown at 40°C. Are all mRNAs affected by the rho protein mutation in the same way? Why or why not?
Problem 22a
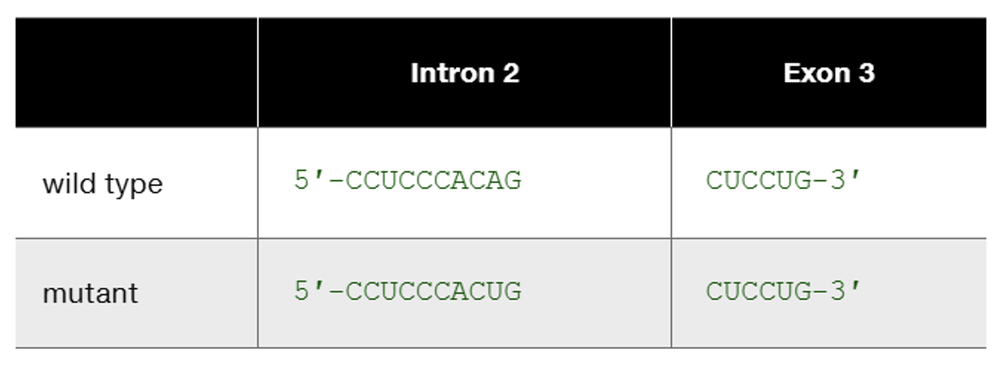
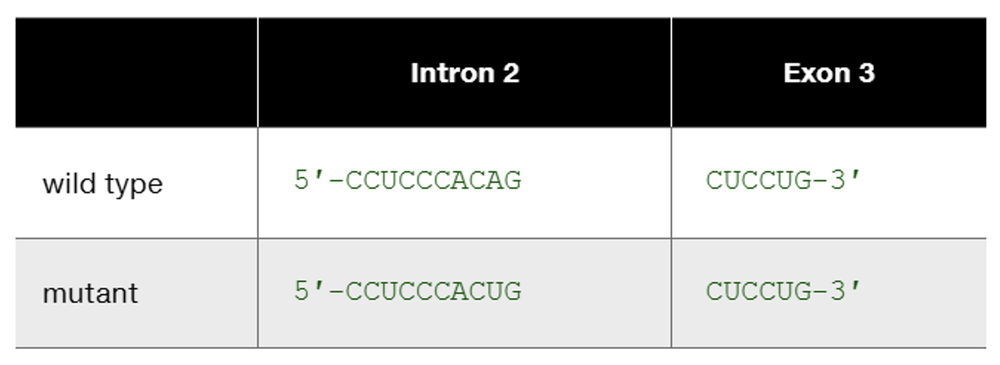
The human β-globin wild-type allele and a certain mutant allele are identical in sequence except for a single base-pair substitution that changes one nucleotide at the end of intron 2. The wild-type and mutant sequences of the affected portion of pre-mRNA are
Speculate about the way in which this base substitution causes mutation of β-globin protein.
Problem 22b
The human β-globin wild-type allele and a certain mutant allele are identical in sequence except for a single base-pair substitution that changes one nucleotide at the end of intron 2. The wild-type and mutant sequences of the affected portion of pre-mRNA are
This is one example of how DNA sequence change occurring somewhere other than in an exon can produce mutation. List other kinds of DNA sequence changes occurring outside exons that can produce mutation. In each case, characterize the kind of change you would expect to see in mutant mRNA or mutant protein.