Back
BackProblem 6b
In Drosophila, the map positions of genes are given in map units numbering from one end of a chromosome to the other. The X chromosome of Drosophila is 66 m.u. long. The X-linked gene for body color—with two alleles, y⁺ for gray body and y for yellow body—resides at one end of the chromosome at map position 0.0. A nearby locus for eye color, with alleles w⁺ for red eye and w for white eye, is located at map position 1.5. A third X-linked gene, controlling bristle form, with f⁺ for normal bristles and f for forked bristles, is located at map position 56.7. At each locus the wild-type allele is dominant over the mutant allele.
Do you expect any of these gene pair(s) to assort independently? Explain your reasoning.
Problem 6c
In Drosophila, the map positions of genes are given in map units numbering from one end of a chromosome to the other. The X chromosome of Drosophila is 66 m.u. long. The X-linked gene for body color—with two alleles, y⁺ for gray body and y for yellow body—resides at one end of the chromosome at map position 0.0. A nearby locus for eye color, with alleles w⁺ for red eye and w for white eye, is located at map position 1.5. A third X-linked gene, controlling bristle form, with f⁺ for normal bristles and f for forked bristles, is located at map position 56.7. At each locus the wild-type allele is dominant over the mutant allele.
A wild-type female fruit fly with the genotype y⁺w⁺f/ywf⁺ is crossed to a male fruit fly that has yellow body, white eye, and forked bristles. Predict the frequency of each progeny phenotype class produced by this mating.
Problem 6d
In Drosophila, the map positions of genes are given in map units numbering from one end of a chromosome to the other. The X chromosome of Drosophila is 66 m.u. long. The X-linked gene for body color—with two alleles, y⁺ for gray body and y for yellow body—resides at one end of the chromosome at map position 0.0. A nearby locus for eye color, with alleles w⁺ for red eye and w for white eye, is located at map position 1.5. A third X-linked gene, controlling bristle form, with f⁺ for normal bristles and f for forked bristles, is located at map position 56.7. At each locus the wild-type allele is dominant over the mutant allele.
Explain how each of the predicted progeny classes is produced.
Problem 7
Genes A, B, and C are linked on a chromosome and found in the order A–B–C. Genes A and B recombine with a frequency of 8%, and genes B and C recombine at a frequency of 24%. For the cross a⁺b⁺c/abc⁺ × abc/abc, predict the frequency of progeny genotypes. Assume interference is zero.
Problem 8a
Gene G recombines with gene T at a frequency of 7%, and gene G recombines with gene R at a frequency of 4%.
Draw two possible genetic maps for these three genes, and identify the recombination frequencies predicted for each map.
Problem 8b
Gene G recombines with gene T at a frequency of 7%, and gene G recombines with gene R at a frequency of 4%.
Assuming that organisms with any desired genotype are available, propose a genetic cross whose result could be used to determine which of the proposed genetic maps is correct.
Problem 9a
Genes A, B, C, D, and E are linked on a chromosome and occur in the order given.
The test cross Ae/aE x ae/ae indicates the genes recombine with a frequency of 28%. If 1000 progeny are produced by this test cross, determine the number of progeny in each outcome class.
Problem 9b
Genes A, B, C, D, and E are linked on a chromosome and occur in the order given.
Previous genetic linkage crosses have determined that recombination frequencies are 6% for genes A and B, 4% for genes B and C, 10% for genes C and D, and 11% for genes D and E. The sum of these frequencies between genes A and E is 31%. Why does the recombination distance between these genes as determined by adding the intervals between adjacent linked genes differ from the distance determined by the test cross?
Problem 10
Syntenic genes can assort independently. Explain this observation.
Problem 11
Define linkage disequilibrium. What is the physical basis of linkage, and what causes linkage equilibrium? Explain how crossing over eliminates linkage disequilibrium.
Problem 12a
On the Drosophila X chromosome, the dominant allele y⁺ produces gray body color and the recessive allele y produces yellow body. This gene is linked to one controlling full eye shape by a dominant allele lz⁺ and lozenge eye shape with a recessive allele lz. These genes recombine with a frequency of approximately 28%. The Lz gene is linked to gene F controlling bristle form, where the dominant phenotype is long bristles and the recessive one is forked bristles. The Lz and F genes recombine with a frequency of approximately 32%.
Using any genotypes you choose, design two separate crosses, one to test recombination between genes Y and Lz and the second between genes Lz and F. Assume 1000 progeny are produced by each cross, and give the number of progeny in each outcome category. (In setting up your crosses, remember that Drosophila males do not undergo recombination.)
Problem 12b
On the Drosophila X chromosome, the dominant allele y⁺ produces gray body color and the recessive allele y produces yellow body. This gene is linked to one controlling full eye shape by a dominant allele lz⁺ and lozenge eye shape with a recessive allele lz. These genes recombine with a frequency of approximately 28%. The Lz gene is linked to gene F controlling bristle form, where the dominant phenotype is long bristles and the recessive one is forked bristles. The Lz and F genes recombine with a frequency of approximately 32%.
Can any cross reveal genetic linkage between gene Y and gene F? Why or why not?
Problem 13a
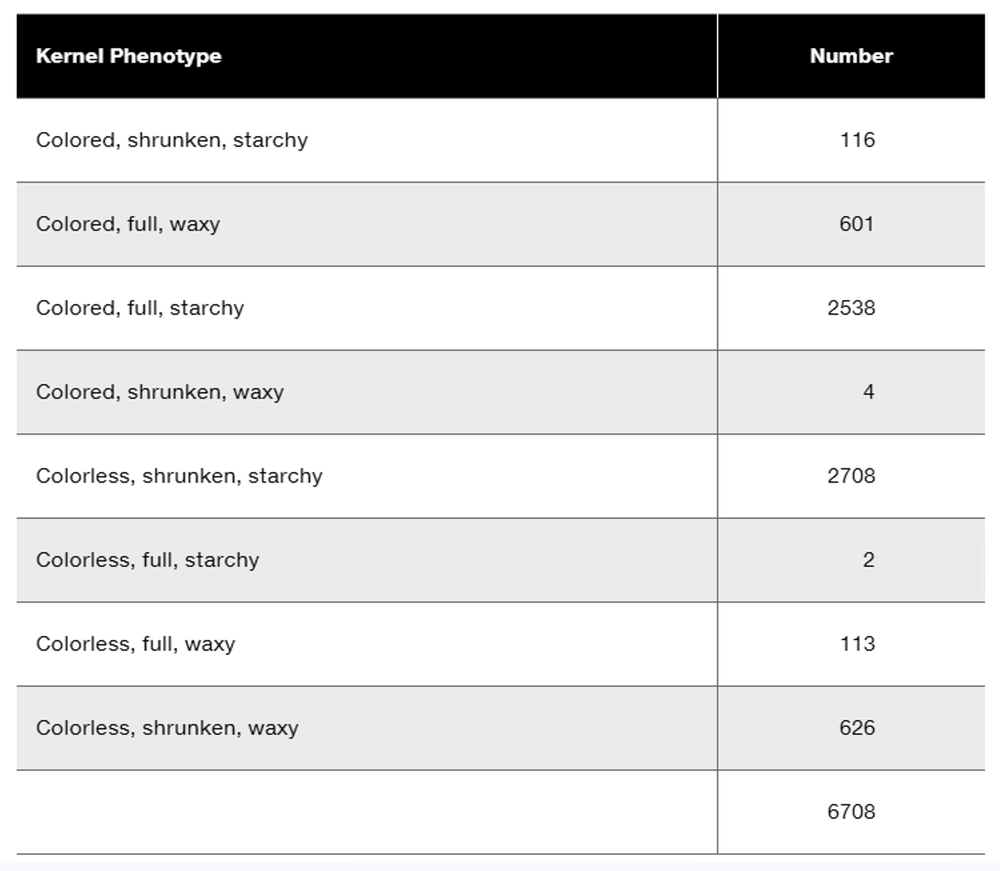
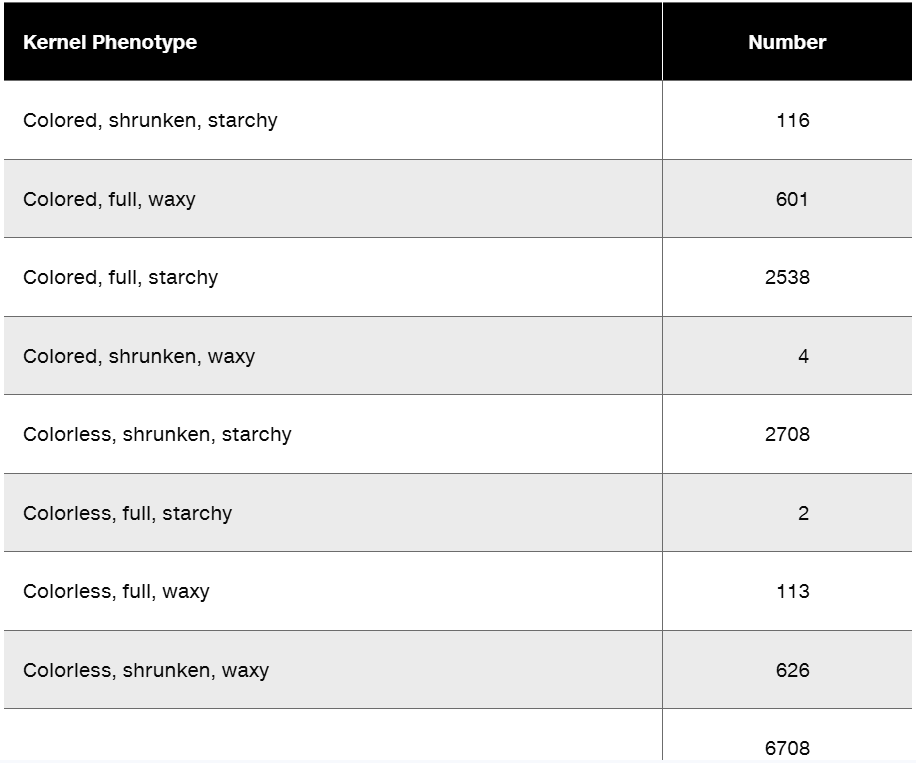
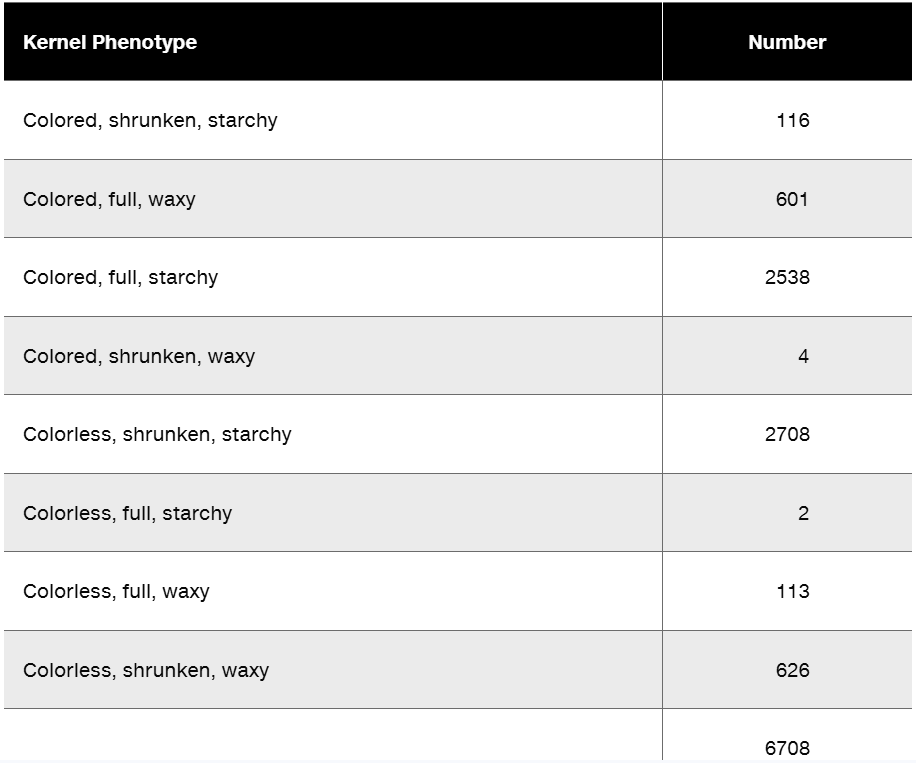
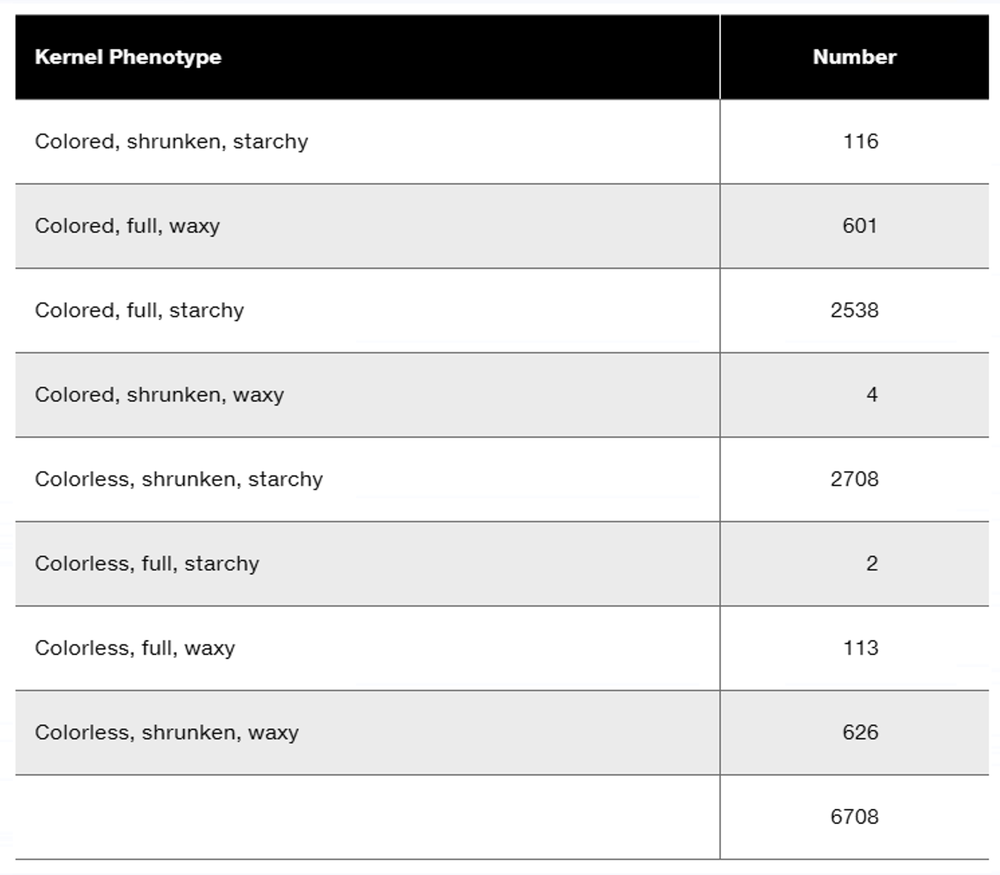
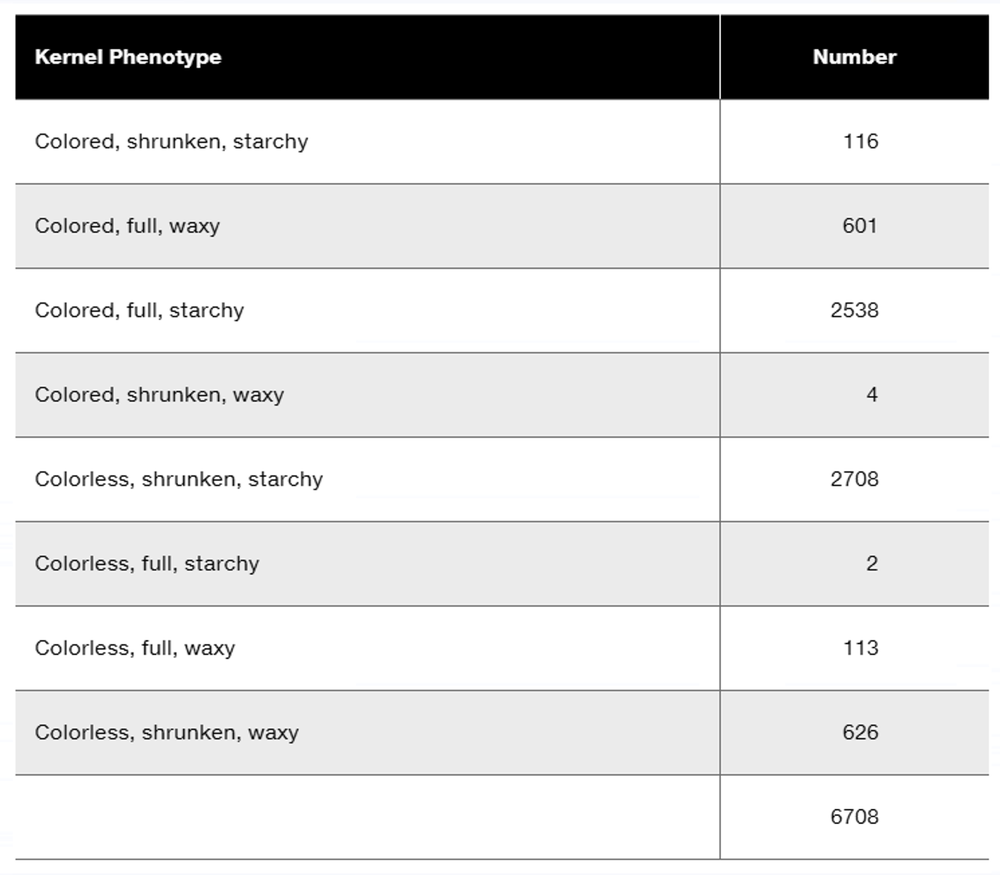
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
Why are these data consistent with genetic linkage among the three genes?
Problem 13b
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
Perform a chi-square test to determine if these data show significant deviation from the expected phenotype distribution.
Problem 13c
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
What is the order of these genes in corn?
Problem 13d
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
Calculate the recombination frequencies between the gene pairs.
Problem 13e
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
What is the interference value for this data set?
Problem 14a
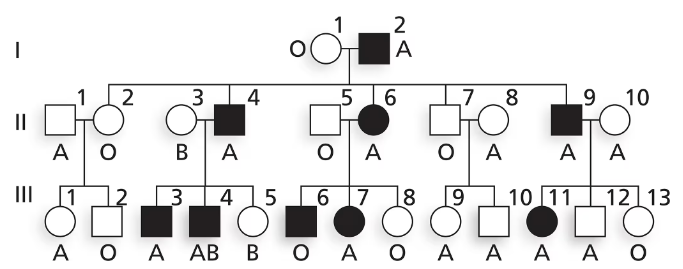
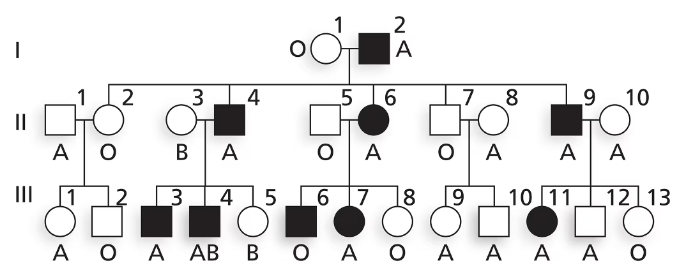
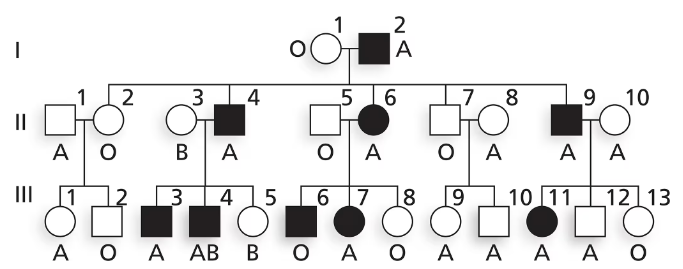
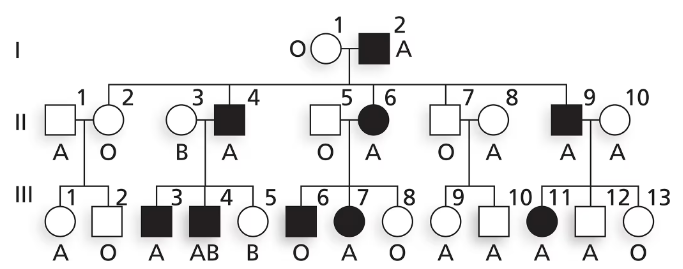
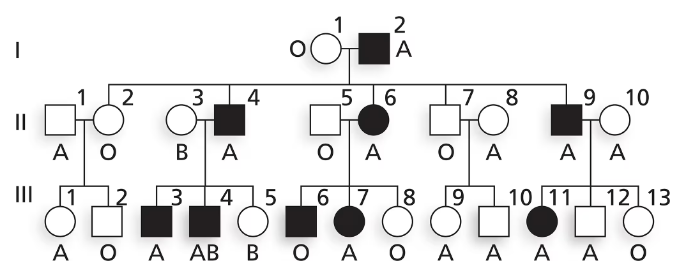
Nail–patella syndrome is an autosomal disorder affecting the shape of nails on fingers and toes as well as the structure of kneecaps. The pedigree below shows the transmission of nail–patella syndrome in a family along with ABO blood type. Is nail–patella syndrome a dominant or a recessive condition?
Is nail–patella syndrome a dominant or a recessive condition? Explain your reasoning.
Problem 14b
Nail–patella syndrome is an autosomal disorder affecting the shape of nails on fingers and toes as well as the structure of kneecaps. The pedigree below shows the transmission of nail–patella syndrome in a family along with ABO blood type.
Does this family give evidence of genetic linkage between nail–patella syndrome and ABO blood group? Why or why not?
Problem 14c
Nail–patella syndrome is an autosomal disorder affecting the shape of nails on fingers and toes as well as the structure of kneecaps. The pedigree below shows the transmission of nail–patella syndrome in a family along with ABO blood type.
Using N and n to represent alleles at the nail–patella locus and Iᴬ, Iᴮ and i to represent ABO alleles, write the genotypes of I-1 and I-2 as well as their five children in generation II.
Problem 14d
Nail–patella syndrome is an autosomal disorder affecting the shape of nails on fingers and toes as well as the structure of kneecaps. The pedigree below shows the transmission of nail–patella syndrome in a family along with ABO blood type.
Explain why III-6 has nail–patella syndrome and III-8 does not. Give genotypes for these two individuals.
Problem 14e
Nail–patella syndrome is an autosomal disorder affecting the shape of nails on fingers and toes as well as the structure of kneecaps. The pedigree below shows the transmission of nail–patella syndrome in a family along with ABO blood type.
Explain why III-11 has nail–patella syndrome and III-12 does not. Give genotypes for these two individuals.
Problem 15a
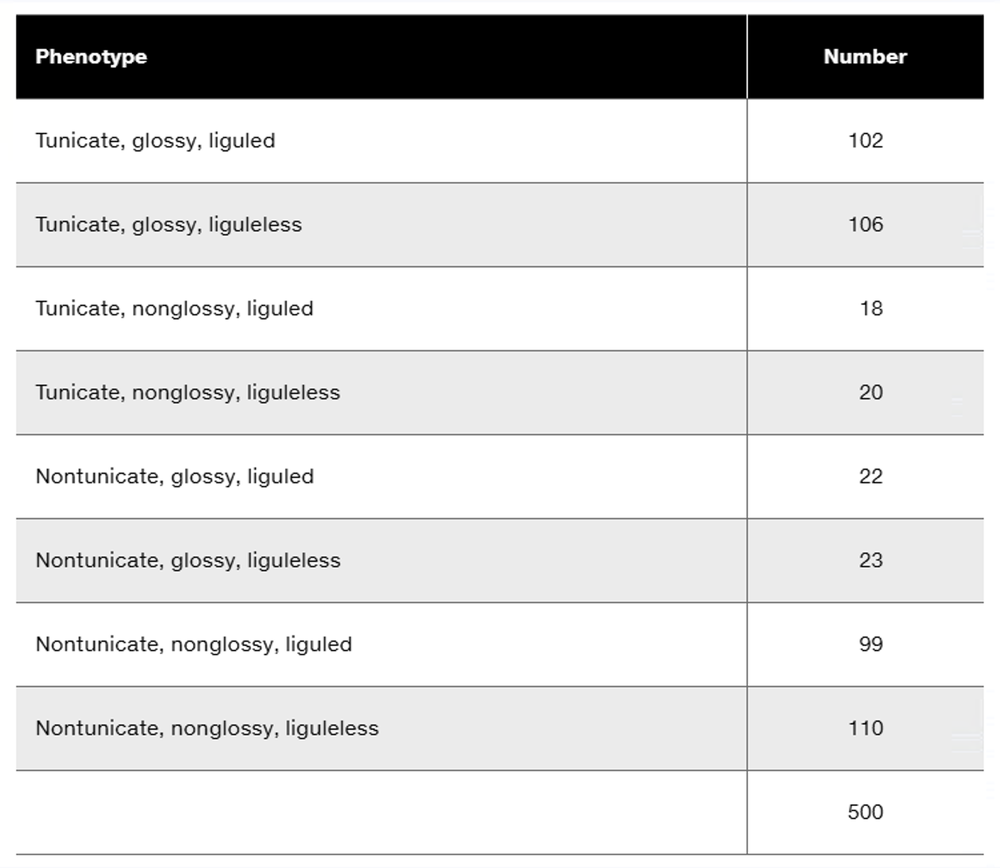
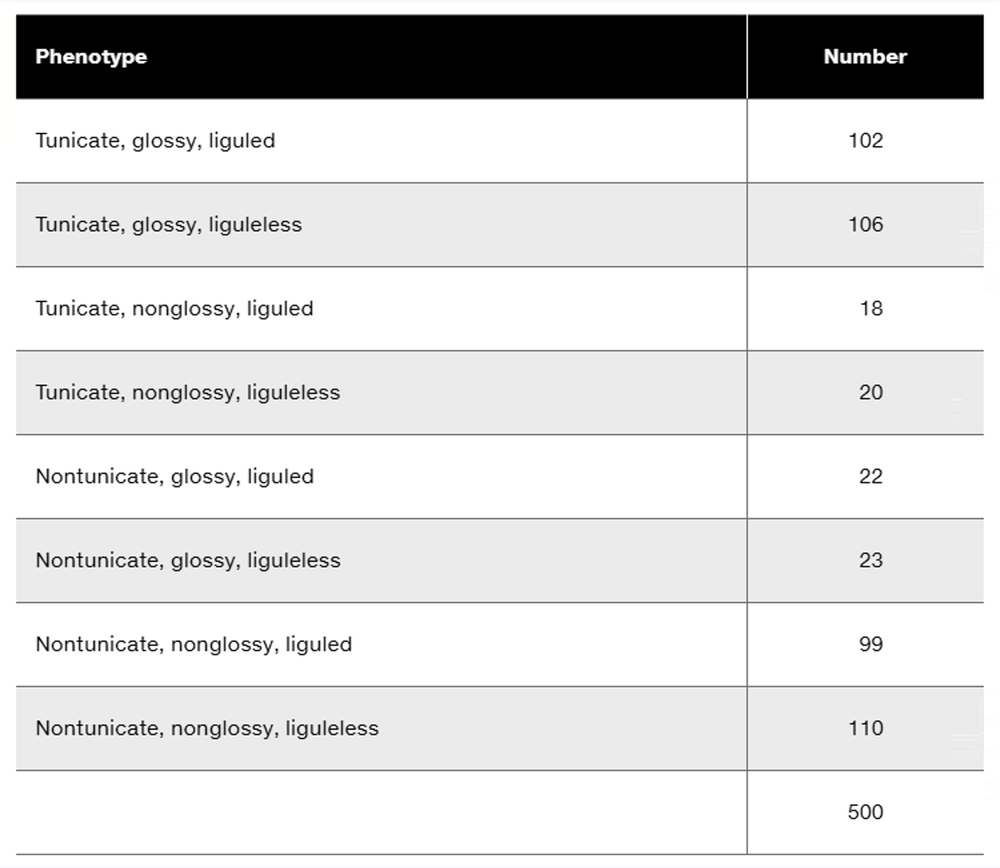
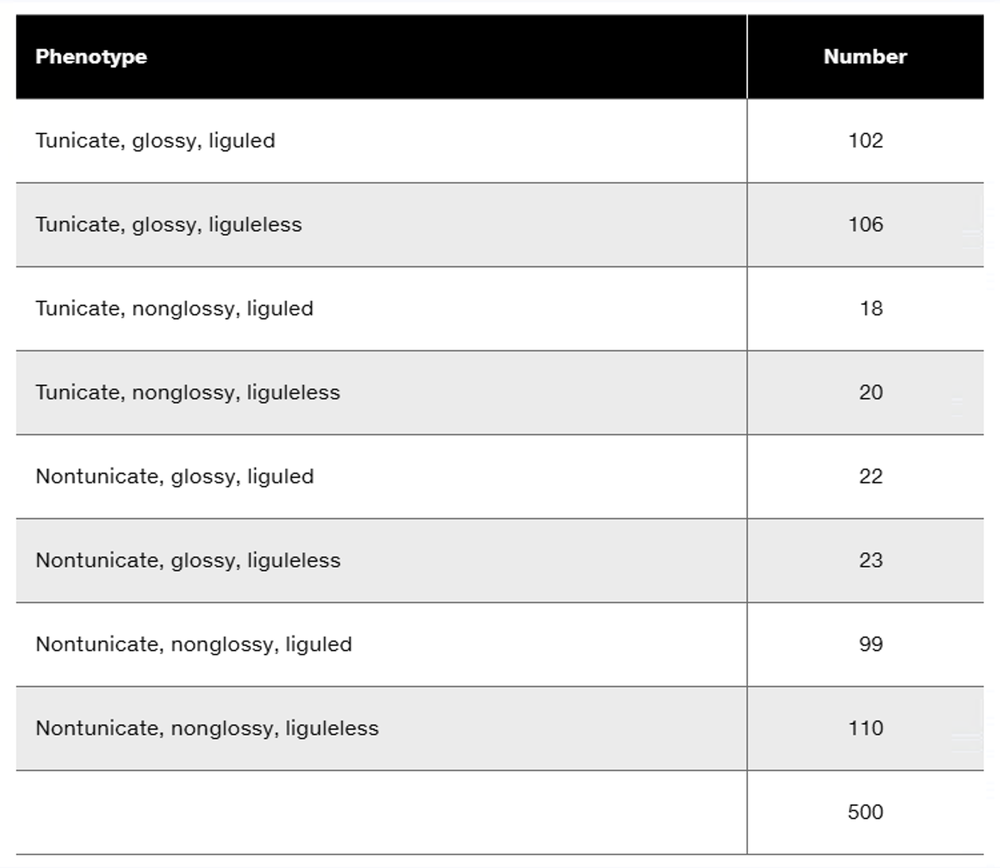
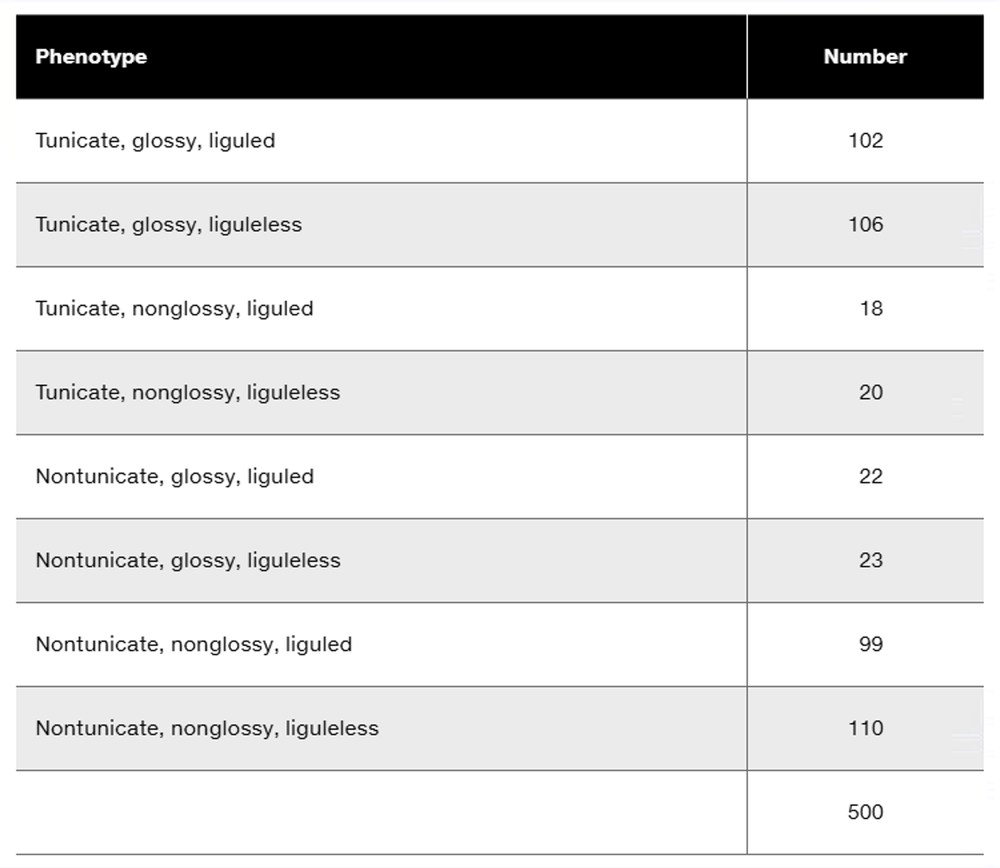
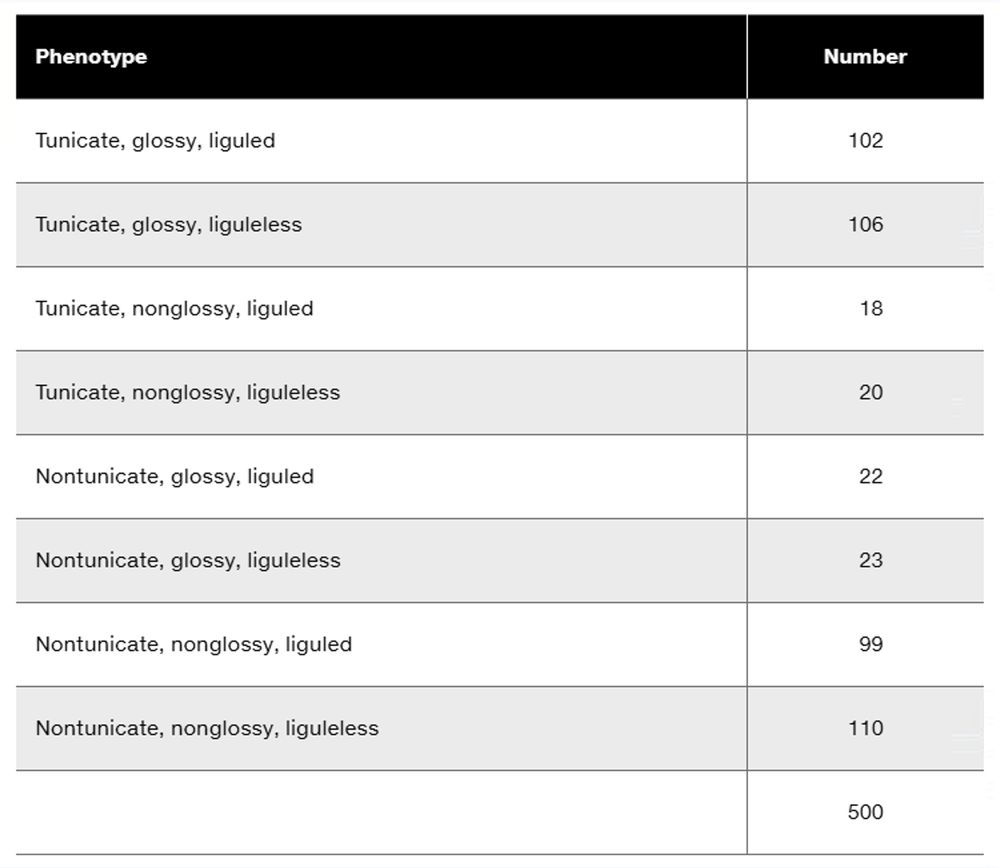
Three dominant traits of corn seedlings, tunicate seed (T-), glossy appearance (G-), and liguled stem (L-), are studied along with their recessive counterparts, nontunicate (tt), nonglossy (gg), and liguleless (ll). A trihybrid plant with the three dominant traits is crossed to a nontunicate, nonglossy, liguleless plant. Kernels on ears of progeny plants are scored for the traits, with the following results:
Is there evidence of genetic linkage among any of these gene pairs? If so, identify the evidence.
Problem 15b
Three dominant traits of corn seedlings, tunicate seed (T-), glossy appearance (G-), and liguled stem (L-), are studied along with their recessive counterparts, nontunicate (tt), nonglossy (gg), and liguleless (ll). A trihybrid plant with the three dominant traits is crossed to a nontunicate, nonglossy, liguleless plant. Kernels on ears of progeny plants are scored for the traits, with the following results:
Is there evidence of independent assortment among any of these gene pairs? If so, identify the evidence.
Problem 15c
Three dominant traits of corn seedlings, tunicate seed (T-), glossy appearance (G-), and liguled stem (L-), are studied along with their recessive counterparts, nontunicate (tt), nonglossy (gg), and liguleless (ll). A trihybrid plant with the three dominant traits is crossed to a nontunicate, nonglossy, liguleless plant. Kernels on ears of progeny plants are scored for the traits, with the following results:
Using the gene symbols given above, write the genotypes of F₁ and F₂ plants.
Problem 15d
Three dominant traits of corn seedlings, tunicate seed (T-), glossy appearance (G-), and liguled stem (L-), are studied along with their recessive counterparts, nontunicate (tt), nonglossy (gg), and liguleless (ll). A trihybrid plant with the three dominant traits is crossed to a nontunicate, nonglossy, liguleless plant. Kernels on ears of progeny plants are scored for the traits, with the following results:
If evidence of linkage is present, calculate the recombination frequency or frequencies from the data presented.
Problem 15e
Three dominant traits of corn seedlings, tunicate seed (T-), glossy appearance (G-), and liguled stem (L-), are studied along with their recessive counterparts, nontunicate (tt), nonglossy (gg), and liguleless (ll). A trihybrid plant with the three dominant traits is crossed to a nontunicate, nonglossy, liguleless plant. Kernels on ears of progeny plants are scored for the traits, with the following results:
Could all three genes be carried on the same chromosome? Discuss why or why not.
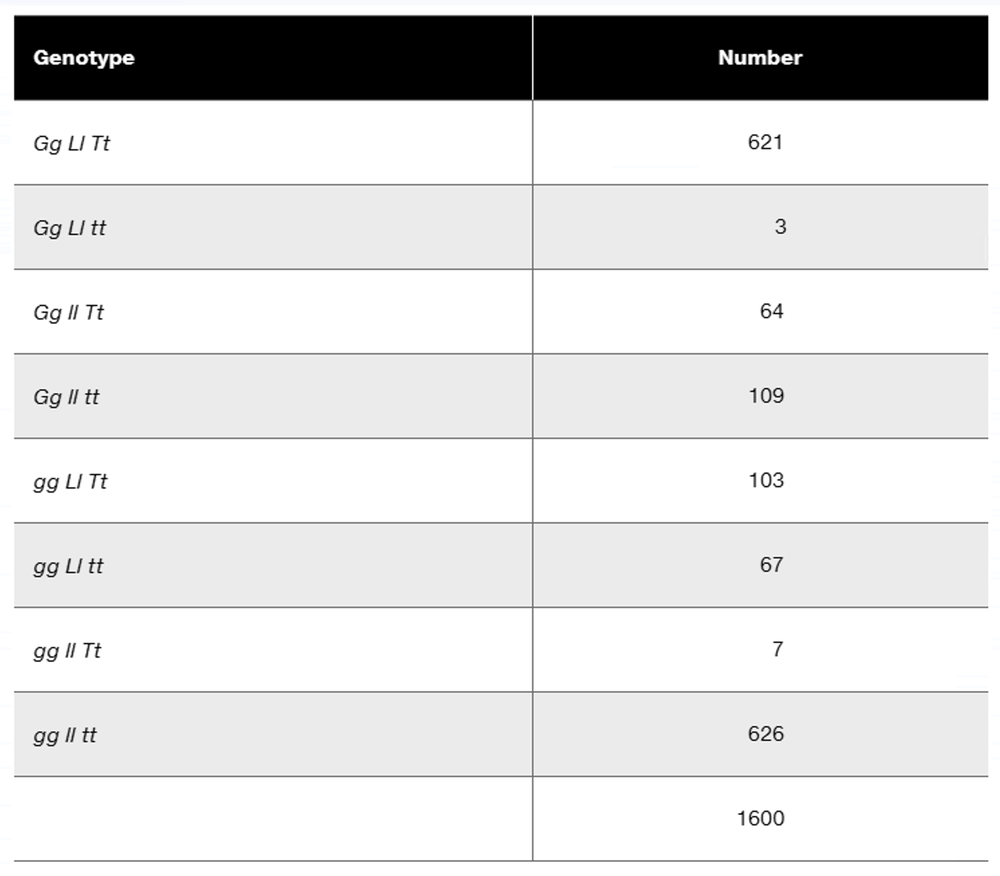
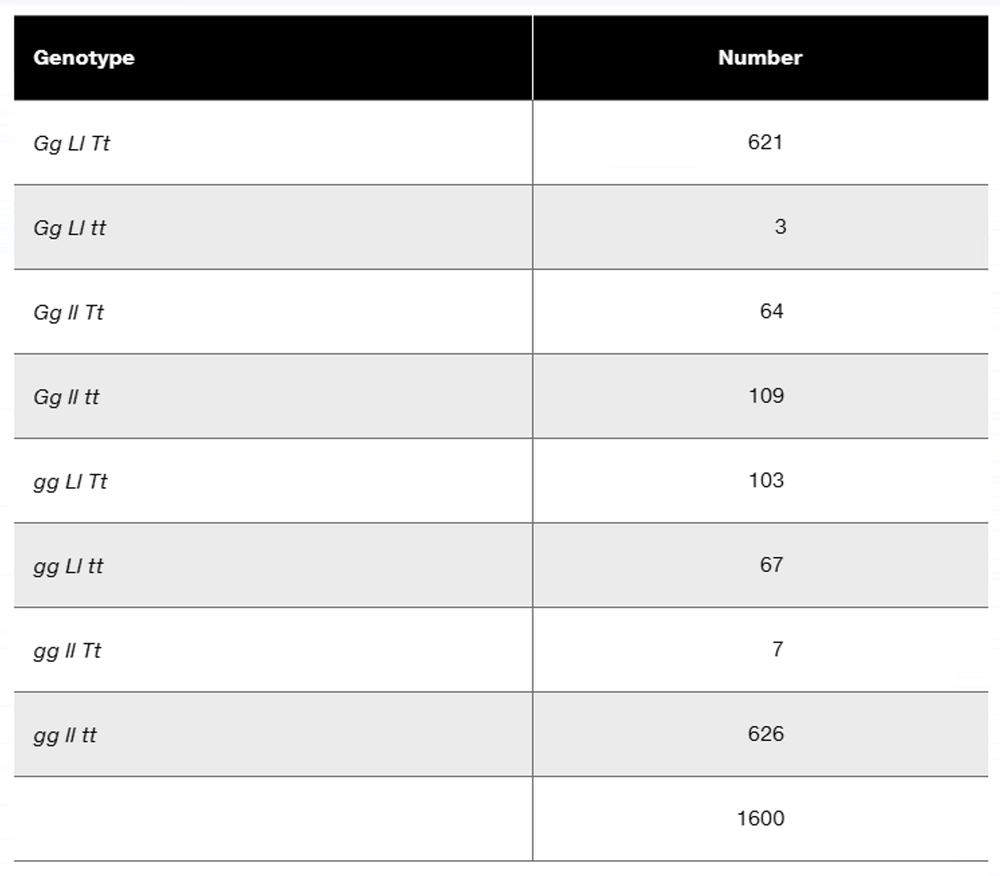
Problem 16a
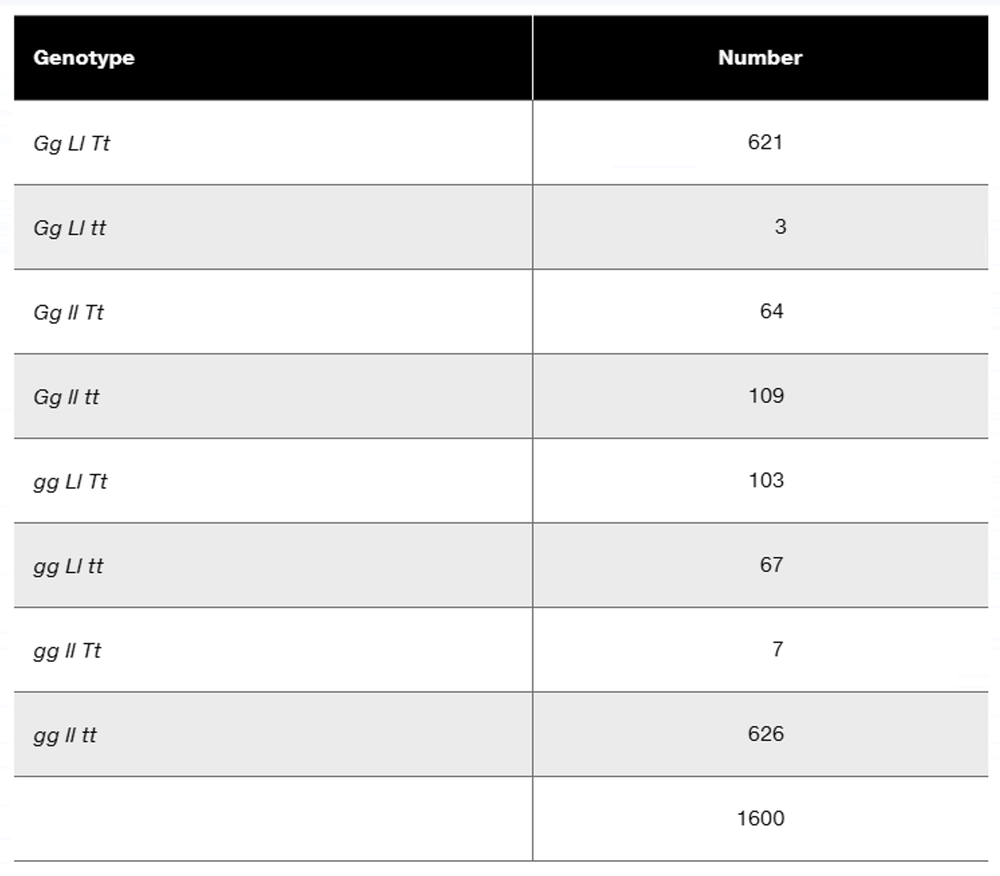
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
What is the order of these three linked genes?
Problem 16b
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
Calculate the recombination frequency between each pair of genes.
Problem 16c
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
Why is the recombination frequency for the outside pair of genes not equal to the sum of recombination frequencies between the adjacent gene pairs?