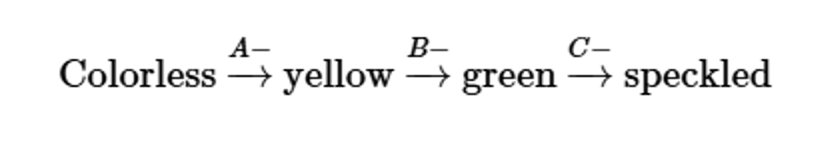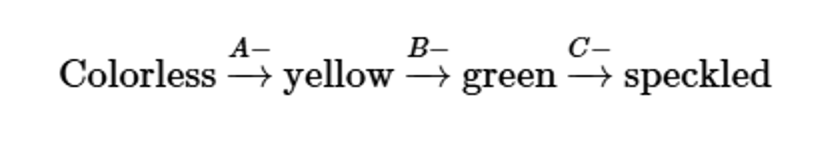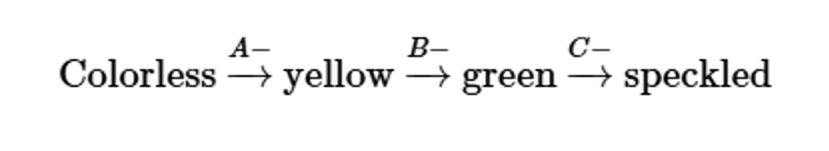Back
BackProblem 26
How do covalent disulfide bonds, hydrogen bonds with water, and hydrophobic interactions all contribute to a protein's tertiary structure?
Problem 27
List as many different categories of protein functions as you can. Wherever possible, give an example of each category.
Problem 28
List three different types of posttranslational modifications that may happen to a protein and the significance of each in the context of protein function.
Problem 29
Why are misfolded proteins a potential problem for the eukaryotic cell, and how do cells combat the accumulation of misfolded proteins?
Problem 30
How does an enzyme function? Why are enzymes essential for living organisms on Earth?
Problem 31
Exon shuffling is a proposal that relates exons in DNA to the repositioning of functional domains in proteins. What evidence exists in support of exon shuffling?
Problem 32a
Three independently assorting genes (A, B, and C) are known to control the following biochemical pathway that provides the basis for flower color in a hypothetical plant:
Three homozygous recessive mutations are also known, each of which interrupts a different one of these steps. Determine the phenotypic results in the F1 and F2 generations resulting from the P1 crosses of true-breeding plants listed here:
speckled (AABBCC) × yellow (AAbbCC)
Problem 32b
Three independently assorting genes (A, B, and C) are known to control the following biochemical pathway that provides the basis for flower color in a hypothetical plant:
Three homozygous recessive mutations are also known, each of which interrupts a different one of these steps. Determine the phenotypic results in the F1 and F2 generations resulting from the P1 crosses of true-breeding plants listed here:
yellow (AAbbCC) × green (AABBcc)
Problem 32c
Three independently assorting genes (A, B, and C) are known to control the following biochemical pathway that provides the basis for flower color in a hypothetical plant:
Three homozygous recessive mutations are also known, each of which interrupts a different one of these steps. Determine the phenotypic results in the F1 and F2 generations resulting from the P1 crosses of true-breeding plants listed here:
colorless (aaBBCC) × green (AABBcc)
Problem 33
How would the results vary in cross (a) of Problem 32 if genes A and B were linked with no crossing over between them? How would the results of cross (a) vary if genes A and B were linked and 20 map units (mu) apart?
Problem 34
Deep in a previously unexplored South American rain forest, a plant species was discovered with true-breeding varieties whose flowers were pink, rose, orange, or purple. A very astute plant geneticist made a single cross, carried to the F₂ generation, as shown:
P₁: purple × pink
F₁: all purple
F₂: 27/64 purple 16/64 pink 12/64 rose 9/64 orange
Based solely on these data, he proposed both a mode of inheritance for flower pigmentation and a biochemical pathway for the synthesis of these pigments. Carefully study the data. Create a hypothesis of your own to explain the mode of inheritance. Then propose a biochemical pathway consistent with your hypothesis. How could you test the hypothesis by making other crosses?
Problem 35
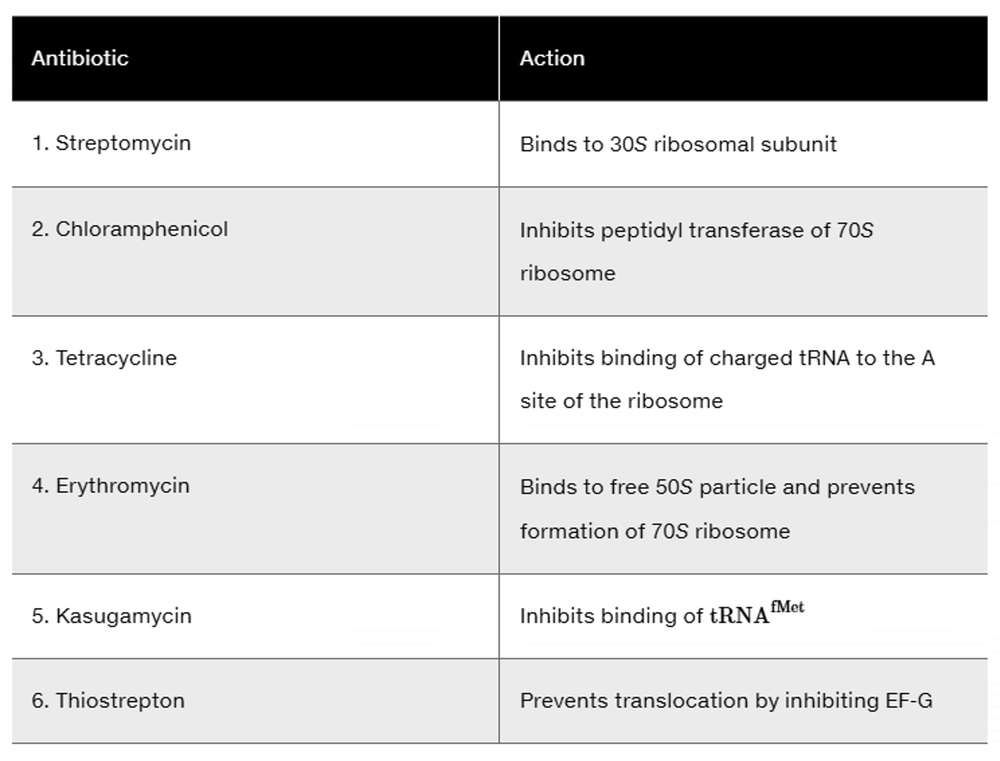
Many antibiotics are effective as drugs to fight off bacterial infections because they inhibit protein synthesis in bacterial cells. Using the information provided in the following table that highlights several antibiotics and their mode of action, discuss which phase of translation is inhibited: initiation, elongation, or termination. What other components of the translational machinery could be targeted to inhibit bacterial protein synthesis?
Problem 36a
The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. Describe how antisense oligonucleotides interrupt the flow of genetic information in a cell.
Problem 36b
The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. What general conclusion can be drawn from the graph?
Problem 36c
The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. What factors other than oligonucleotide length are likely to influence antisense efficacy in vivo?
Problem 37a
Infantile cardiomyopathy is a devastating disorder that is fatal during the first year of life due to defects in the function of heart muscles resulting from mitochondrial dysfunction. A study, performed by Götz et al. [(2011). Am. J. Hum. Genet. 88:635–642), identified two different causative mutations in the gene for mitochondrial alanyl-tRNA synthetase (mtAlaRS). One mutation changes a leucine residue at amino acid position 155 to arginine (p.Leu155Arg). The other mutation changes arginine at position 592 to tryptophan (p.Arg592Trp). The mtAlaRS enzyme has an N-terminal domain (amino acids 36–481) that catalyzes tRNA aminoacylation and an internal editing domain (amino acids 484–782) that catalyzes deacylation in the case that the tRNA is charged with the wrong amino acid.
Consider the position of the disease causing missense mutations in the mtAlaRS gene in the context of the known protein domains of this enzyme. What predictions can you make about how these mutations impair protein synthesis within mitochondria in different ways?
Problem 37b
Infantile cardiomyopathy is a devastating disorder that is fatal during the first year of life due to defects in the function of heart muscles resulting from mitochondrial dysfunction. A study, performed by Götz et al. [(2011). Am. J. Hum. Genet. 88:635–642), identified two different causative mutations in the gene for mitochondrial alanyl-tRNA synthetase (mtAlaRS). One mutation changes a leucine residue at amino acid position 155 to arginine (p.Leu155Arg). The other mutation changes arginine at position 592 to tryptophan (p.Arg592Trp). The mtAlaRS enzyme has an N-terminal domain (amino acids 36–481) that catalyzes tRNA aminoacylation and an internal editing domain (amino acids 484–782) that catalyzes deacylation in the case that the tRNA is charged with the wrong amino acid.
Which mutation would you predict has a more severe impairment of translation in mitochondria, and why?