Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
4. Genetic Mapping and Linkage
Trihybrid Cross
Struggling with Genetics?
Join thousands of students who trust us to help them ace their exams!Watch the first videoMultiple Choice
The following table shows data from a cross examining three genes (a, b, and c). Calculate the recombination frequency for A and C
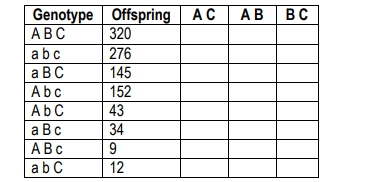
A
32%
B
37%
C
20%
D
9.8%
0 Comments
 Verified step by step guidance
Verified step by step guidance1
Identify the parental and recombinant genotypes from the table. Parental genotypes are those with the highest number of offspring, typically the first two rows: 'A B C' and 'a b c'.
Determine the recombinant genotypes for genes A and C. These are the genotypes where A and C are not inherited together as in the parental types. From the table, these are 'A b C', 'a B c', 'A B c', and 'a b C'.
Sum the number of offspring for the recombinant genotypes identified in the previous step. This involves adding the numbers for 'A b C', 'a B c', 'A B c', and 'a b C'.
Calculate the total number of offspring by summing all the values in the 'Offspring' column of the table.
Compute the recombination frequency using the formula: \( \text{Recombination Frequency} = \frac{\text{Number of Recombinant Offspring}}{\text{Total Number of Offspring}} \times 100 \). This will give you the percentage of recombination between genes A and C.
Related Videos
Related Practice
Open Question
Drosophila melanogaster has one pair of sex chromosomes (XX or XY) and three pairs of autosomes, referred to as chromosomes II, III, and IV. A genetics student discovered a male fly with very short (sh) legs. Using this male, the student was able to establish a pure breeding stock of this mutant and found that it was recessive. She then incorporated the mutant into a stock containing the recessive gene black (b, body color located on chromosome II) and the recessive gene pink (p, eye color located on chromosome III). A female from the homozygous black, pink, short stock was then mated to a wild-type male. The F₁ males of this cross were all wild type and were then backcrossed to the homozygous b, p, sh females. The F₂ results appeared as shown in the following table. No other phenotypes were observed.Wild Pink* Black, Black, Pink,Short* ShortFemales 63 58 55 69Males 59 65 51 60*Other trait or traits are wild type.The student repeated the experiment, making the reciprocal cross, F₁ females backcrossed to homozygous b, p, sh males. She observed that 85 percent of the offspring fell into the given classes, but that 15 percent of the offspring were equally divided among b + p, b + +, + sh p, and + sh + phenotypic males and females. How can these results be explained, and what information can be derived from the data?
Trihybrid Cross practice set




