In analyzing the number of different bases in a DNA sample, which result would be consistent with the base-pairing rules?
a. A=G
b. A+G=C+T
c. A+T=G+C
d. A=C

 Verified step by step guidance
Verified step by step guidance
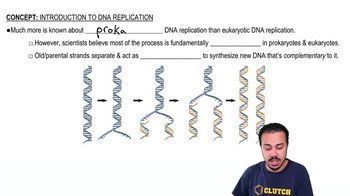
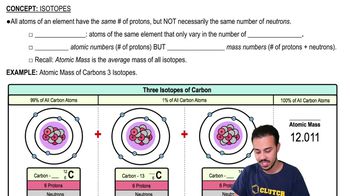
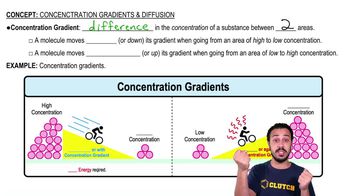
In analyzing the number of different bases in a DNA sample, which result would be consistent with the base-pairing rules?
a. A=G
b. A+G=C+T
c. A+T=G+C
d. A=C
The elongation of the leading strand during DNA synthesis
a. Progresses away from the replication fork
b. Occurs in the 3′→5′ direction
c. Produces Okazaki fragments
d. Depends on the action of DNA polymerase
In a nucleosome, the DNA is wrapped around
a. Histones
b. Ribosomes
c. Polymerase molecules
d. A thymine dimer
A biochemist isolates, purifies, and combines in a test tube a variety of molecules needed for DNA replication. When she adds some DNA to the mixture, replication occurs, but each DNA molecule consists of a normal strand paired with numerous segments of DNA a few hundred nucleotides long. What has she probably left out of the mixture?
a. DNA polymerase
b. DNA ligase
c. Okazaki fragments
d. Primase
The spontaneous loss of amino groups from adenine in DNA results in hypoxanthine, an uncommon base, opposite thymine. What combination of proteins could repair such damage?
a. Nuclease, DNA polymerase, DNA ligase
b. Telomerase, primase, DNA polymerase
c. Telomerase, helicase, single-strand binding protein
d. DNA ligase, replication fork proteins, adenylyl cyclase
Although the proteins that cause the E. coli chromosome to coil are not histones, what property would you expect them to share with histones, given their ability to bind to DNA (see Figure 5.14)?